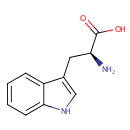
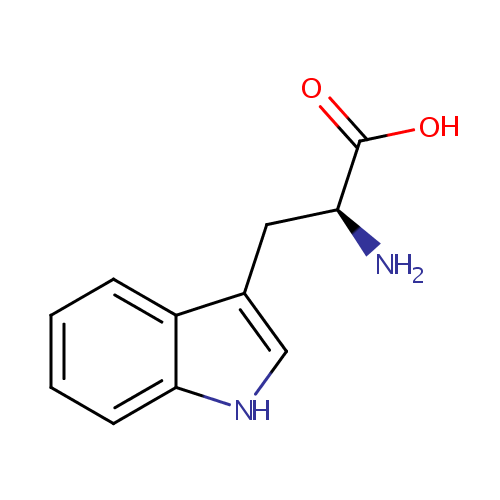
L-Tryptophan (PAMDB000203)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000203 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | L-Tryptophan | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | Tryptophan is an amino acid which is the precursor of serotonin. Plants and microorganisms commonly synthesize tryptophan from shikimic acid or anthranilate. The latter condenses with phosphoribosylpyrophosphate (PRPP), generating pyrophosphate as a by-product. After ring opening of the ribose moiety and following reductive decarboxylation, indole-3-glycerinephosphate is produced, which in turn is transformed into indole. In the last step, tryptophan synthase catalyzes the formation of tryptophan from indole and the amino acid, serine. Metabolism of tryptophan to serotonin requires nutrients such as vitamin B6, niacin and glutathione. Niacin is an important metabolite of tryptophan. (Wikipedia) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C11H12N2O2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 204.2252 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 204.089877638 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | QIVBCDIJIAJPQS-VIFPVBQESA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C11H12N2O2/c12-9(11(14)15)5-7-6-13-10-4-2-1-3-8(7)10/h1-4,6,9,13H,5,12H2,(H,14,15)/t9-/m0/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 73-22-3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | (2S)-2-amino-3-(1H-indol-3-yl)propanoic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | L-tryptophan | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | N[C@@H](CC1=CNC2=CC=CC=C12)C(O)=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as indolyl carboxylic acids and derivatives. These are compounds containing a carboxylic acid chain (of at least 2 carbon atoms) linked to an indole ring. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organoheterocyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Indoles and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Indolyl carboxylic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Indolyl carboxylic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic heteropolycyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | 0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | 230 °C | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Indole + L-Serine > Water + L-Tryptophan Indoleglycerol phosphate + L-Serine > D-Glyceraldehyde 3-phosphate + Water + L-Tryptophan N-Methyltryptophan + Water + Oxygen > Formaldehyde + Hydrogen peroxide + L-Tryptophan Adenosine triphosphate + tRNA(Trp) + L-Tryptophan + tRNA(Trp) <> Adenosine monophosphate + Pyrophosphate + L-Tryptophanyl-tRNA(Trp) Water + L-Tryptophan <> Indole + Ammonium + Pyruvic acid L-Tryptophan + Water <> Indole + Pyruvic acid + Ammonia Adenosine triphosphate + L-Tryptophan + tRNA(Trp) <> Adenosine monophosphate + Pyrophosphate + L-Tryptophanyl-tRNA(Trp) L-Tryptophan + Water <> Hydrogen ion + Indole + Pyruvic acid + Ammonia Adenosine triphosphate + L-Tryptophan + tRNA(Trp) > Adenosine monophosphate + Pyrophosphate + L-tryptophyl-tRNA(Trp) L-Serine + Indoleglycerol phosphate > L-Tryptophan + glyceraldehyde 3-phosphate + Water L-Tryptophan + Water + 2-Aminoacrylic acid + 2-Iminopropanoate <> Indole + Pyruvic acid + Ammonia L-Serine + Indoleglycerol phosphate + Indole <> L-Tryptophan + D-Glyceraldehyde 3-phosphate + Water L-Tryptophan + Adenosine triphosphate + Hydrogen ion + tRNA(Trp) > Adenosine monophosphate + Pyrophosphate + L-tryptophyl-tRNA(Trp) Indole + L-Serine + L-Serine > Water + L-Tryptophan L-Tryptophan > Hydrogen ion + Indole + 2-Aminoacrylic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Amir-Heidari, Bagher; Thirlway, Jenny; Micklefield, Jason. Stereochemical course of tryptophan dehydrogenation during biosynthesis of the calcium-dependent lipopeptide antibiotics. Organic Letters (2007), 9(8), 1513-1516. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Download (PDF) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in nucleotide binding
- Specific function:
- ATP + L-tryptophan + tRNA(Trp) = AMP + diphosphate + L-tryptophyl-tRNA(Trp)
- Gene Name:
- trpS
- Locus Tag:
- PA4439
- Molecular weight:
- 49 kDa
Reactions
| ATP + L-tryptophan + tRNA(Trp) = AMP + diphosphate + L-tryptophyl-tRNA(Trp). |
- General function:
- Involved in catalytic activity
- Specific function:
- The alpha subunit is responsible for the aldol cleavage of indoleglycerol phosphate to indole and glyceraldehyde 3- phosphate
- Gene Name:
- trpA
- Locus Tag:
- PA0035
- Molecular weight:
- 28.5 kDa
Reactions
| L-serine + 1-C-(indol-3-yl)glycerol 3-phosphate = L-tryptophan + glyceraldehyde 3-phosphate + H(2)O. |
- General function:
- Involved in catalytic activity
- Specific function:
- The beta subunit is responsible for the synthesis of L- tryptophan from indole and L-serine
- Gene Name:
- trpB
- Locus Tag:
- PA0036
- Molecular weight:
- 43.7 kDa
Reactions
| L-serine + 1-C-(indol-3-yl)glycerol 3-phosphate = L-tryptophan + glyceraldehyde 3-phosphate + H(2)O. |
Transporters
- General function:
- Involved in nucleotide binding
- Specific function:
- Probably part of a binding-protein-dependent transport system yecCS for an amino acid. Probably responsible for energy coupling to the transport system
- Gene Name:
- yecC
- Locus Tag:
- PA5152
- Molecular weight:
- 28.4 kDa
- General function:
- Involved in amino acid transmembrane transporter activity
- Specific function:
- Involved in transporting tryptophan across the cytoplasmic membrane
- Gene Name:
- mtr
- Locus Tag:
- PA5434
- Molecular weight:
- 44.7 kDa
- General function:
- Involved in transport
- Specific function:
- Permease that is involved in the transport across the cytoplasmic membrane of the aromatic amino acids (phenylalanine, tyrosine, and tryptophan)
- Gene Name:
- aroP
- Locus Tag:
- PA3000
- Molecular weight:
- 51 kDa