FADH2 (PAMDB000293)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000293 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
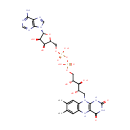
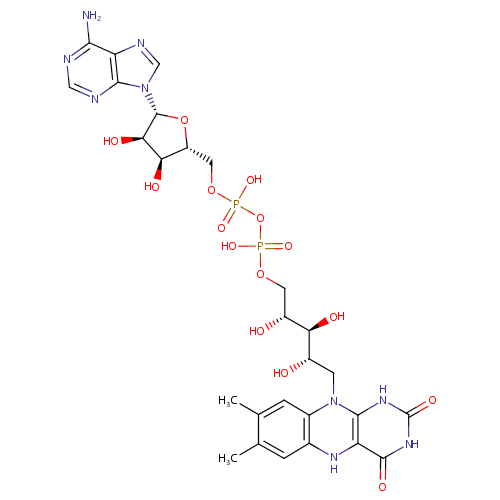
| Name: | FADH2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | FADH2 is a reduced form of Flavin adenine dinucleotide (FAD). FAD is a redox cofactor involved in several important reactions in metabolism. It can exist in two different redox states, which it converts between by accepting or donating electrons. The molecule consists of a riboflavin moiety (vitamin B2) bound to the phosphate group of an ADP molecule. The flavin group is bound to ribitol, a sugar alcohol, by a carbon-nitrogen bond, not a glycosidic bond. Thus, riboflavin is not technically a nucleotide; the name flavin adenine dinucleotide is a misnomer. FAD can be reduced to FADH2, whereby it accepts two hydrogen atoms. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C27H35N9O15P2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 787.5656 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 787.172784519 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | YPZRHBJKEMOYQH-UYBVJOGSSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C27H35N9O15P2/c1-10-3-12-13(4-11(10)2)35(24-18(32-12)25(42)34-27(43)33-24)5-14(37)19(39)15(38)6-48-52(44,45)51-53(46,47)49-7-16-20(40)21(41)26(50-16)36-9-31-17-22(28)29-8-30-23(17)36/h3-4,8-9,14-16,19-21,26,32,37-41H,5-7H2,1-2H3,(H,44,45)(H,46,47)(H2,28,29,30)(H2,33,34,42,43)/t14-,15+,16+,19-,20+,21+,26+/m0/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 1910-41-4 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | [({[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy]({[(2R,3S,4S)-5-{7,8-dimethyl-2,4-dioxo-1H,2H,3H,4H,5H,10H-benzo[g]pteridin-10-yl}-2,3,4-trihydroxypentyl]oxy})phosphinic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | fadh(.) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | CC1=CC2=C(C=C1C)N(C[C@H](O)[C@H](O)[C@H](O)COP(O)(=O)OP(O)(=O)OC[C@H]1O[C@H]([C@H](O)[C@@H]1O)N1C=NC3=C1N=CN=C3N)C1=C(N2)C(=O)NC(=O)N1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as flavin nucleotides. These are nucleotides containing a flavin moiety. Flavin is a compound that contains the tricyclic isoalloxazine ring system, which bears 2 oxo groups at the 2- and 4-positions. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Nucleosides, nucleotides, and analogues | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Flavin nucleotides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Flavin nucleotides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic heteropolycyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | FADH2 + 2 Hydrogen ion + SufBCD with two bound [2Fe-2S] clusters > FAD + SufBCD with bound [4Fe-4S] cluster FAD + Hydrogen ion + NADPH > FADH2 + NADP Adenosine triphosphate + FADH2 + 2 Iron + Water + SufBCD scaffold complex + 2 SufSE with bound sulfur > ADP + FAD +7 Hydrogen ion + Phosphate + SufBCD with bound [2Fe-2S] cluster +2 SufSE sulfur acceptor complex Adenosine triphosphate + FADH2 + 2 Iron + Water + SufBCD with bound [2Fe-2S] cluster + 2 SufSE with bound sulfur > ADP + FAD +7 Hydrogen ion + Phosphate + SufBCD with two bound [2Fe-2S] clusters +2 SufSE sulfur acceptor complex FADH2 + 2 Iron + 2 IscS with bound sulfur + IscU scaffold protein > FAD +6 Hydrogen ion +2 IscS sulfur acceptor protein + IscU with bound [2Fe-2S] cluster FADH2 + 2 Iron + 2 IscS with bound sulfur + IscU with bound [2Fe-2S] cluster > FAD +6 Hydrogen ion +2 IscS sulfur acceptor protein + IscU with two bound [2Fe-2S] clusters Butyryl-CoA + FAD <> Crotonoyl-CoA + FADH2 FAD + Octanoyl-CoA <> FADH2 + (2E)-Octenoyl-CoA FAD + Palmityl-CoA <> FADH2 + (2E)-Hexadecenoyl-CoA FAD + Tetradecanoyl-CoA <> FADH2 + (2E)-Tetradecenoyl-CoA FAD + Hexanoyl-CoA <> FADH2 + trans-2-Hexenoyl-CoA FAD + Stearoyl-CoA <> FADH2 + Trans-Octadec-2-enoyl-CoA Lauroyl-CoA + FAD <> (2E)-Dodecenoyl-CoA + FADH2 Decanoyl-CoA (N-C10:0CoA) + FAD <> (2E)-Decenoyl-CoA + FADH2 FAD + L-Proline > L-D-1-Pyrroline-5-carboxylic acid + FADH2 + Hydrogen ion D-Alanine + FAD + Water > FADH2 + Ammonium + Pyruvic acid FADH2 + 2 Hydrogen ion + IscU with two bound [2Fe-2S] clusters > FAD + IscU with bound [4Fe-4S] cluster FADH2 + 2 Fe3+ > FAD +2 Iron +2 Hydrogen ion FAD + Hydrogen ion + NADH > FADH2 + NAD FADH2 + 2 Ferroxamine > FAD +2 Iron +2 ferroxamine minus Fe(3) +2 Hydrogen ion Succinic acid + FAD <> FADH2 + Fumaric acid Glycerol 3-phosphate + FAD <> Dihydroxyacetone phosphate + FADH2 Butanoyl-CoA + FAD <> FADH2 + Crotonoyl-CoA L-Malic acid + FAD <> FADH2 + Oxalacetic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Kavakli I Halil; Sancar Aziz Analysis of the role of intraprotein electron transfer in photoreactivation by DNA photolyase in vivo. Biochemistry (2004), 43(48), 15103-10. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in electron carrier activity
- Specific function:
- Two distinct, membrane-bound, FAD-containing enzymes are responsible for the catalysis of fumarate and succinate interconversion; the fumarate reductase is used in anaerobic growth, and the succinate dehydrogenase is used in aerobic growth
- Gene Name:
- sdhB
- Locus Tag:
- PA1584
- Molecular weight:
- 26.2 kDa
Reactions
| Succinate + acceptor = fumarate + reduced acceptor. |
- General function:
- Involved in oxidoreductase activity
- Specific function:
- Oxidizes proline to glutamate for use as a carbon and nitrogen source and also function as a transcriptional repressor of the put operon
- Gene Name:
- putA
- Locus Tag:
- PA0782
- Molecular weight:
- 115.6 kDa
Reactions
| L-proline + acceptor = (S)-1-pyrroline-5-carboxylate + reduced acceptor. |
| (S)-1-pyrroline-5-carboxylate + NAD(P)(+) + 2 H(2)O = L-glutamate + NAD(P)H. |
- General function:
- Involved in metabolic process
- Specific function:
- Catalyzes the removal of elemental sulfur and selenium atoms from cysteine and selenocysteine to produce alanine. Functions as a sulfur delivery protein for NAD, biotin and Fe-S cluster synthesis. Transfers sulfur on 'Cys-456' of thiI in a transpersulfidation reaction. Transfers sulfur on 'Cys-19' of tusA in a transpersulfidation reaction. Functions also as a selenium delivery protein in the pathway for the biosynthesis of selenophosphate
- Gene Name:
- iscS
- Locus Tag:
- PA3814
- Molecular weight:
- 44.7 kDa
Reactions
| L-cysteine + acceptor = L-alanine + S-sulfanyl-acceptor. |
- General function:
- Involved in D-amino-acid dehydrogenase activity
- Specific function:
- Oxidative deamination of D-amino acids
- Gene Name:
- dadA
- Locus Tag:
- PA5304
- Molecular weight:
- 47.1 kDa
Reactions
| A D-amino acid + H(2)O + acceptor = a 2-oxo acid + NH(3) + reduced acceptor. |
- General function:
- Involved in electron carrier activity
- Specific function:
- Two distinct, membrane-bound, FAD-containing enzymes are responsible for the catalysis of fumarate and succinate interconversion; the fumarate reductase is used in anaerobic growth, and the succinate dehydrogenase is used in aerobic growth
- Gene Name:
- sdhA
- Locus Tag:
- PA1583
- Molecular weight:
- 63.5 kDa
Reactions
| Succinate + acceptor = fumarate + reduced acceptor. |
- General function:
- Involved in succinate dehydrogenase activity
- Specific function:
- Membrane-anchoring subunit of succinate dehydrogenase (SDH)
- Gene Name:
- sdhD
- Locus Tag:
- PA1582
- Molecular weight:
- 13.7 kDa
- General function:
- Involved in oxidoreductase activity
- Specific function:
- Conversion of glycerol 3-phosphate to dihydroxyacetone. Uses molecular oxygen or nitrate as electron acceptor
- Gene Name:
- glpD
- Locus Tag:
- PA3584
- Molecular weight:
- 57.1 kDa
Reactions
| sn-glycerol 3-phosphate + a quinone = glycerone phosphate + a quinol. |
- General function:
- Involved in sulfite reductase (NADPH) activity
- Specific function:
- Component of the sulfite reductase complex that catalyzes the 6-electron reduction of sulfite to sulfide. This is one of several activities required for the biosynthesis of L- cysteine from sulfate
- Gene Name:
- cysI
- Locus Tag:
- PA1838
- Molecular weight:
- 62.1 kDa
Reactions
| H(2)S + 3 NADP(+) + 3 H(2)O = sulfite + 3 NADPH. |
- General function:
- Involved in malate dehydrogenase (quinone) activity
- Specific function:
- (S)-malate + a quinone = oxaloacetate + reduced quinone
- Gene Name:
- mqo
- Locus Tag:
- PA3452
- Molecular weight:
- 57.2 kDa
Reactions
| (S)-malate + a quinone = oxaloacetate + reduced quinone. |
- General function:
- Involved in succinate dehydrogenase activity
- Specific function:
- Membrane-anchoring subunit of succinate dehydrogenase (SDH)
- Gene Name:
- sdhC
- Locus Tag:
- PA1581
- Molecular weight:
- 13.7 kDa
- General function:
- Involved in acyl-CoA dehydrogenase activity
- Specific function:
- Catalyzes the dehydrogenation of acyl-CoA
- Gene Name:
- fadE
- Locus Tag:
- PA2815
- Molecular weight:
- 88.8 kDa
Reactions
| An acyl-CoA + FAD = a dehydrogenated acyl-CoA + FADH(2). |
- General function:
- Involved in iron ion binding
- Specific function:
- May be involved in the formation or repair of [Fe-S] clusters present in iron-sulfur proteins (Potential)
- Gene Name:
- nifU
- Locus Tag:
- PA3813
- Molecular weight:
- 13.8 kDa