Search Results for compounds
Searching compounds for
returned 4373 results.
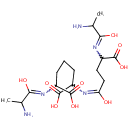
L-alanine-D-glutamate-meso-2,6-diaminoheptanedioate (PAMDB001648)
IUPAC:
2-[(2-amino-1-hydroxypropylidene)amino]-6-({4-[(2-amino-1-hydroxypropylidene)amino]-4-carboxy-1-hydroxybutylidene}amino)heptanedioic acid
CAS: Not Available
Description: L-alanine-d-glutamate-meso-2,6-diaminoheptanedioate belongs to the class of Peptides. These are compounds containing an amide derived from two or more amino carboxylic acid molecules (the same or different) by formation of a covalent bond from the carbonyl carbon of one to the nitrogen atom of another. (inferred from compound structure)
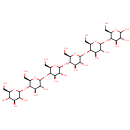
Maltohexaose (PAMDB001651)
IUPAC:
(2R,3R,4S,5S,6R)-2-{[(2R,3S,4R,5R,6R)-6-{[(2R,3S,4R,5R,6R)-6-{[(2R,3S,4R,5R,6R)-6-{[(2R,3S,4R,5R,6R)-4,5-dihydroxy-2-(hydroxymethyl)-6-{[(2R,3S,4R,5R)-4,5,6-trihydroxy-2-(hydroxymethyl)oxan-3-yl]oxy}oxan-3-yl]oxy}-4,5-dihydroxy-2-(hydroxymethyl)oxan-3-yl]oxy}-4,5-dihydroxy-2-(hydroxymethyl)oxan-3-yl]oxy}-4,5-dihydroxy-2-(hydroxymethyl)oxan-3-yl]oxy}-6-(hydroxymethyl)oxane-3,4,5-triol
CAS: 34620-77-4
Description: Maltohexaose is hexasaccaride or more specifically a hexasaccharide comprised of six D-glucose residues connected by alpha(1->4) linkages. It is a substrate for energy metabolism and carbon for Pseudomonas aeruginosa. Maltohexaose is imported into the cell via the maltooligosaccharide-specific LamB-channel of Pseudomonas aeruginosa (also called maltoporin). It is degraded by alpha amylase.
Maltopentaose (PAMDB001652)
IUPAC:
6-({6-[(6-{[4,5-dihydroxy-2-(hydroxymethyl)-6-[2,3,4-trihydroxy-1-(hydroxymethoxy)butoxy]oxan-3-yl]oxy}-2,4,5-trihydroxyoxan-3-yl)oxy]-4,5-dihydroxy-2-(hydroxymethyl)oxan-3-yl}oxy)oxane-2,3,4,5-tetrol
CAS: 34620-76-3
Description: Maltopentaose is pentasaccaride or more specifically a pentasaccharide comprised of five D-glucose residues connected by alpha(1->4) linkages. It is a substrate for energy metabolism and carbon for Pseudomonas aeruginosa. Maltopentaose is imported into the cell via the maltooligosaccharide-specific LamB-channel of Pseudomonas aeruginosa (also called maltoporin). It is degraded by alpha amylase.
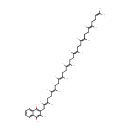
Menaquinol 8 (PAMDB001653)
IUPAC:
2-methyl-3-[(2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-octamethyldotriaconta-2,6,10,14,18,22,26,30-octaen-1-yl]naphthalene-1,4-diol
CAS: Not Available
Description: Menaquinol 8 is a polyprenylhydroquinone having a an octaprenyl moiety at position 2 and a methyl group at position 3. It is a substrate for Dimethyl sulfoxide reductase (dmsA). This enzyme catalyzes the reduction of dimethyl sulfoxide (DMSO) to dimethyl sulfide (DMS) using the following reaction: Dimethylsulfide + menaquinone + H2O = dimethylsulfoxide + menaquinol. DMSO reductase serves as the terminal reductase under anaerobic conditions, with DMSO being the terminal electron acceptor. Terminal reductase during anaerobic growth on various sulfoxides and N-oxide compounds. This enzyme allows P. aeruginosa to grow anaerobically on DMSO as respiratory oxidant. Menaquinol 8 is generated by Ubiquinone/menaquinone biosynthesis methyltransferase (ubiE). This enzyme is required for the conversion of demethylmenaquinone (DMKH2) to menaquinone (MKH2) and has the following catalytic activity: A demethylmenaquinone + S-adenosyl-L-methionine = a menaquinol + S-adenosyl-L-homocysteine.
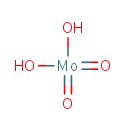
Molybdate (PAMDB001654)
IUPAC:
dioxomolybdenumdiol
CAS: 14259-85-9
Description: Molybdate is a compound containing an oxoanion with molybdenum in its highest oxidation state of 6. Molybdenum can form a very large range of such oxoanions which can be discrete structures or polymeric extended structures, although the latter are only found in the solid state.The larger oxoanions are members of group of compounds termed polyoxometalates, and because they contain only one type of metal atom are often called isopolymetalates. The discrete molybdenum oxoanions range in size from the simplest MoO42_, found in potassium molybdate up to extremely large structures found in isopoly-molybdenum blues that contain for example 154 Mo atoms. The behaviour of molybdenum is different from the other elements in group 6. Molybdate is involved in the molybdenum cofactor biosynthesis pathway. Molybdate reacts with molybdopterin-AMP to produce molybdenum cofactor, AMP, and H2O.
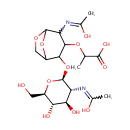
N-Acetyl-D-glucosamine(anhydrous)N-Acetylmuramic acid (PAMDB001656)
IUPAC:
2-[(2-{[(2S,3R,4R,5S,6R)-4,5-dihydroxy-3-[(1-hydroxyethylidene)amino]-6-(hydroxymethyl)oxan-2-yl]oxy}-4-[(1-hydroxyethylidene)amino]-6,8-dioxabicyclo[3.2.1]octan-3-yl)oxy]propanoic acid
CAS: Not Available
Description: N-acetyl-d-glucosamine(anhydrous)n-acetylmuramic acid belongs to the class of Hexoses. These are monosaccharides in which the sugar unit is a hexose. (inferred from compound structure)
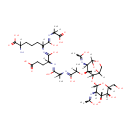
N-Acetyl-D-glucosamine(anhydrous)N-Acetylmuramyl-tetrapeptide (PAMDB001657)
IUPAC:
2-amino-6-{[4-carboxy-2-({2-[(2-{[(3R,4R)-2-{[(2S,3R,4R,5S,6R)-4,5-dihydroxy-3-[(1-hydroxyethylidene)amino]-6-(hydroxymethyl)oxan-2-yl]oxy}-4-[(1-hydroxyethylidene)amino]-6,8-dioxabicyclo[3.2.1]octan-3-yl]oxy}-1-hydroxypropylidene)amino]-1-hydroxypropylidene}amino)-1-hydroxybutylidene]amino}-6-[(1-carboxyethyl)-C-hydroxycarbonimidoyl]hexanoic acid
CAS: Not Available
Description: N-acetyl-D-glucosamine(anhydrous)N-acetylmuramyl-tetrapeptide is an intermediate in peptidoglycan synthesis and turnover. Peptidoglycan can be described as a fisherman's net that encloses bacteria. The mesh of the net is made of two segments of parallel, somewhat inextensible glycan threads, held together by two small elastic peptide crosslinks allowing the net to expand or shrink. The glycan moiety of the peptidoglycan is very uniform among all bacteria, and is made up of alternating β-1,4-linked N-acetylglucosamine and N-acetyl muramate residues, with an average chain lengthof 10 to 65 disaccharide units (depending on the organism). The peptidoglycan synthesis pathway starts in the cytoplasm, where in six steps the peptidoglycan precursor a UDP-N-acetylmuramoyl-pentapeptide is synthesized. This precursor is then attached to the memberane acceptor all-trans-undecaprenyl phosphate, generating a N-acetylmuramoyl-pentapeptide-diphosphoundecaprenol, also known as lipid I. Another transferase then adds UDP-N-acetyl-α-D-glucosamine, yielding the complete monomeric unit a lipid II, also known as lipid II. This final lipid intermediate is transferred by an as yet unknown mechanism through the membrane. The peptidoglycan monomers are then polymerized on the outside surface by glycosyltransferases, which form the linear glycan chains, and transpeptidases, which catalyze the formation of peptide crosslinks. Peptide crosslinks form between different parts of the peptides depending on the organism. For example, in Mycobacteria and in Pseudomonas aeruginosa most links form between the carboxyl group of the penultimate D-alanine (residue 4) of one peptide to the amino group at the D-center of meso-diaminopimelate (residue 3) of an adjacent peptide of a second glycan chain (as in Pseudomonas aeruginosa). The crosslinking reaction is catalyzed by transpeptidases and involves the cleavage of the D-alanyl-D-alanine bond of the donor peptide, providing the energy to drive the reaction. As a result, the peptides in the peptidoglycan polymers are one or two amino acids shorter than the peptides in the monomers.
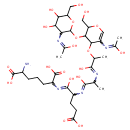
N-Acetyl-D-glucosamine(anhydrous)N-Acetylmuramyl-tripeptide (PAMDB001658)
IUPAC:
2-amino-6-{[4-carboxy-2-({2-[(2-{[3-({4,5-dihydroxy-3-[(1-hydroxyethylidene)amino]-6-(hydroxymethyl)oxan-2-yl}oxy)-5-[(1-hydroxyethylidene)amino]-2-(hydroxymethyl)-3,4-dihydro-2H-pyran-4-yl]oxy}-1-hydroxypropylidene)amino]-1-hydroxypropylidene}amino)-1-hydroxybutylidene]amino}heptanedioic acid
CAS: Not Available
Description: N-acetyl-D-glucosamine(anhydrous)N-acetylmuramyl-tripeptide is an intermediate in peptidoglycan synthesis and turnover. Peptidoglycan can be described as a fisherman's net that encloses bacteria. The mesh of the net is made of two segments of parallel, somewhat inextensible glycan threads, held together by two small elastic peptide crosslinks allowing the net to expand or shrink. The glycan moiety of the peptidoglycan is very uniform among all bacteria, and is made up of alternating β-1,4-linked N-acetylglucosamine and N-acetyl muramate residues, with an average chain lengthof 10 to 65 disaccharide units (depending on the organism). The peptidoglycan synthesis pathway starts in the cytoplasm, where in six steps the peptidoglycan precursor a UDP-N-acetylmuramoyl-pentapeptide is synthesized. This precursor is then attached to the memberane acceptor all-trans-undecaprenyl phosphate, generating a N-acetylmuramoyl-pentapeptide-diphosphoundecaprenol, also known as lipid I. Another transferase then adds UDP-N-acetyl-α-D-glucosamine, yielding the complete monomeric unit a lipid II, also known as lipid II. This final lipid intermediate is transferred by an as yet unknown mechanism through the membrane. The peptidoglycan monomers are then polymerized on the outside surface by glycosyltransferases, which form the linear glycan chains, and transpeptidases, which catalyze the formation of peptide crosslinks. Peptide crosslinks form between different parts of the peptides depending on the organism. For example, in Mycobacteria and in Pseudomonas aeruginosa most links form between the carboxyl group of the penultimate D-alanine (residue 4) of one peptide to the amino group at the D-center of meso-diaminopimelate (residue 3) of an adjacent peptide of a second glycan chain (as in Pseudomonas aeruginosa). The crosslinking reaction is catalyzed by transpeptidases and involves the cleavage of the D-alanyl-D-alanine bond of the donor peptide, providing the energy to drive the reaction. As a result, the peptides in the peptidoglycan polymers are one or two amino acids shorter than the peptides in the monomers.
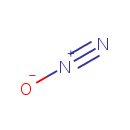
Nitrous oxide (PAMDB001663)
IUPAC:
diazooxidane
CAS: 10024-97-2
Description: Nitrous oxide, commonly known as laughing gas, is a chemical compound with the formula N2O. It is an oxide of nitrogen. At room temperature, it is a colorless, non-flammable gas, with a slightly sweet odor and taste. N2O is produced naturally by microorganisms in soil through various types of nitrification and denitrification processes. (Wikipedia) Nitrous oxide production by Pseudomonas aeruginosa seems to result from the reduction of nitrite (NO2-) by nitrate (NO3-) reductase. (PMID 6347062)
Octadecanoyl-phosphate (n-C18:0) (PAMDB001666)
IUPAC:
(octadecanoyloxy)phosphonate
CAS: Not Available
Description: Octadecanoyl-phosphate (n-c18:0) belongs to the class of Acyl Phosphates. These are organic compounds containing the functional group -CO-P(O)(O)OH. (inferred from compound structure)