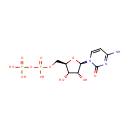
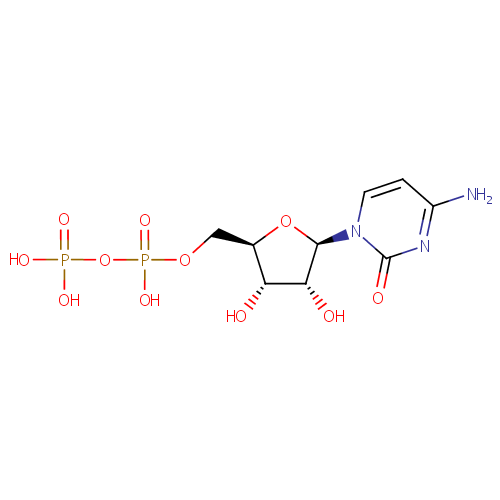
| References: |
- Carstensen S, Pliska-Matyshak G, Bhuvarahamurthy N, Robbins KM, Murthy PP: Biosynthesis and localization of phosphatidyl-scyllo-inositol in barley aleurone cells. Lipids. 1999 Jan;34(1):67-73. Pubmed: 10188599
- George TP, Cook HW, Byers DM, Palmer FB, Spence MW: Inhibition of phosphatidylcholine and phosphatidylethanolamine biosynthesis by cytochalasin B in cultured glioma cells: potential regulation of biosynthesis by Ca(2+)-dependent mechanisms. Biochim Biophys Acta. 1991 Jul 9;1084(2):185-93. Pubmed: 1854804
- George TP, Morash SC, Cook HW, Byers DM, Palmer FB, Spence MW: Phosphatidylcholine biosynthesis in cultured glioma cells: evidence for channeling of intermediates. Biochim Biophys Acta. 1989 Aug 22;1004(3):283-91. Pubmed: 2758024
- Houtkooper RH, Akbari H, van Lenthe H, Kulik W, Wanders RJ, Frentzen M, Vaz FM: Identification and characterization of human cardiolipin synthase. FEBS Lett. 2006 May 29;580(13):3059-64. Epub 2006 Apr 27. Pubmed: 16678169
- Houtkooper RH, Vaz FM: Cardiolipin, the heart of mitochondrial metabolism. Cell Mol Life Sci. 2008 Aug;65(16):2493-506. doi: 10.1007/s00018-008-8030-5. Pubmed: 18425414
- Ishii, N., Nakahigashi, K., Baba, T., Robert, M., Soga, T., Kanai, A., Hirasawa, T., Naba, M., Hirai, K., Hoque, A., Ho, P. Y., Kakazu, Y., Sugawara, K., Igarashi, S., Harada, S., Masuda, T., Sugiyama, N., Togashi, T., Hasegawa, M., Takai, Y., Yugi, K., Arakawa, K., Iwata, N., Toya, Y., Nakayama, Y., Nishioka, T., Shimizu, K., Mori, H., Tomita, M. (2007). "Multiple high-throughput analyses monitor the response of E. coli to perturbations." Science 316:593-597. Pubmed: 17379776
- Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
- Keseler, I. M., Collado-Vides, J., Santos-Zavaleta, A., Peralta-Gil, M., Gama-Castro, S., Muniz-Rascado, L., Bonavides-Martinez, C., Paley, S., Krummenacker, M., Altman, T., Kaipa, P., Spaulding, A., Pacheco, J., Latendresse, M., Fulcher, C., Sarker, M., Shearer, A. G., Mackie, A., Paulsen, I., Gunsalus, R. P., Karp, P. D. (2011). "EcoCyc: a comprehensive database of Escherichia coli biology." Nucleic Acids Res 39:D583-D590. Pubmed: 21097882
- Lindblad L, Schersten T: Incorporation rate in vitro of choline and methyl-methionine into human hepatic lecithins. Scand J Gastroenterol. 1976;11(6):587-91. Pubmed: 981963
- Nowicki M, Muller F, Frentzen M: Cardiolipin synthase of Arabidopsis thaliana. FEBS Lett. 2005 Apr 11;579(10):2161-5. Pubmed: 15811335
- Riekhof WR, Voelker DR: Uptake and utilization of lyso-phosphatidylethanolamine by Saccharomyces cerevisiae. J Biol Chem. 2006 Dec 1;281(48):36588-96. Epub 2006 Oct 2. Pubmed: 17015438
- Tsitolovskii LE, Kraevskii AA: [Possible relation between learning and non-template RNA synthesis in neurons]. Zh Vyssh Nerv Deiat Im I P Pavlova. 1982 Mar-Apr;32(2):284-91. Pubmed: 6178232
- van der Werf, M. J., Overkamp, K. M., Muilwijk, B., Coulier, L., Hankemeier, T. (2007). "Microbial metabolomics: toward a platform with full metabolome coverage." Anal Biochem 370:17-25. Pubmed: 17765195
- Winder, C. L., Dunn, W. B., Schuler, S., Broadhurst, D., Jarvis, R., Stephens, G. M., Goodacre, R. (2008). "Global metabolic profiling of Escherichia coli cultures: an evaluation of methods for quenching and extraction of intracellular metabolites." Anal Chem 80:2939-2948. Pubmed: 18331064
|
|---|