Search Results for compounds
Searching compounds for
returned 4373 results.
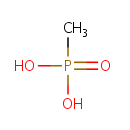
Methylphosphonate (PAMDB001160)
IUPAC:
methylphosphonic acid
CAS: 993-13-5
Description: Methylphosphonate is a member of the chemical class known as Organic Phosphonic Acids and Derivatives. These are organic compounds containing phosphonic acid or a derivative thereof. . Phosphonates (Pn) are a large class of organophosphorus molecules that have direct carbon-phosphorus (C-P) bonds in place of the carbon-oxygen-phosphorus ester bond. In bacteria two pathways exist for Pn breakdown for use as a P source: the phosphonatase and C-P lyase pathways. These pathways differ both in regard to their substrate specificity and their cleavage mechanism. The phosphonatase pathway acts on the natural Pn alpha-aminoethylphosphonate (AEPn). In a two-step process it leads to cleavage of the C-P bond by a hydrolysis reaction requiring an adjacent carbonyl group. In contrast the C-P lyase pathway has a broad substrate specificity. It leads to cleavage of substituted Pn (such as AEPn) as well as unsubstituted Pn by a mechanism involving redox or radical chemistry. Due to its broad substrate specificity, the C-P lyase pathway is generally thought to be responsible for the breakdown of Pn herbicides (such as glyphosate) by bacteria.
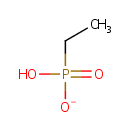
Ethylphosphonate (PAMDB001161)
IUPAC:
hydrogen ethylphosphonate
CAS: 6779-09-5
Description: Ethylphosphonate is a member of the chemical class known as Organic Phosphonic Acids and Derivatives. These are organic compounds containing phosphonic acid or a derivative thereof. phosphonoacetaldehyde is catalyzed by phosphonoacetaldehyde hydrolase. This is the first nucleic sequence report of the phosphonoacetaldehyde hydrolase, an enzyme involved in the carbon-phosphorus bond cleavage. (PMID 9332393). Phosphonates (Pn) are a large class of organophosphorus molecules that have direct carbon-phosphorus (C-P) bonds in place of the carbon-oxygen-phosphorus ester bond. In bacteria two pathways exist for Pn breakdown for use as a P source: the phosphonatase and C-P lyase pathways. These pathways differ both in regard to their substrate specificity and their cleavage mechanism. The phosphonatase pathway acts on the natural Pn alpha-aminoethylphosphonate (AEPn). In a two-step process it leads to cleavage of the C-P bond by a hydrolysis reaction requiring an adjacent carbonyl group. In contrast the C-P lyase pathway has a broad substrate specificity. It leads to cleavage of substituted Pn (such as AEPn) as well as unsubstituted Pn by a mechanism involving redox or radical chemistry. Due to its broad substrate specificity, the C-P lyase pathway is generally thought to be responsible for the breakdown of Pn herbicides (such as glyphosate) by bacteria.
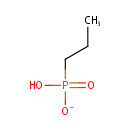
Propylphosphonate (PAMDB001162)
IUPAC:
hydrogen propylphosphonate
CAS: Not Available
Description: Propylphosphonate is a member of the chemical class known as Organic Phosphonic Acids and Derivatives. These are organic compounds containing phosphonic acid or a derivative thereof. . Phosphonates (Pn) are a large class of organophosphorus molecules that have direct carbon-phosphorus (C-P) bonds in place of the carbon-oxygen-phosphorus ester bond. In bacteria two pathways exist for Pn breakdown for use as a P source: the phosphonatase and C-P lyase pathways. These pathways differ both in regard to their substrate specificity and their cleavage mechanism. The phosphonatase pathway acts on the natural Pn alpha-aminoethylphosphonate (AEPn). In a two-step process it leads to cleavage of the C-P bond by a hydrolysis reaction requiring an adjacent carbonyl group. In contrast the C-P lyase pathway has a broad substrate specificity. It leads to cleavage of substituted Pn (such as AEPn) as well as unsubstituted Pn by a mechanism involving redox or radical chemistry. Due to its broad substrate specificity, the C-P lyase pathway is generally thought to be responsible for the breakdown of Pn herbicides (such as glyphosate) by bacteria.
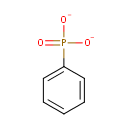
Phenylphosphonate (PAMDB001164)
IUPAC:
phenylphosphonate
CAS: 1571-33-1
Description: Phenylphosphonate is a member of the chemical class known as Phenylphosphines and Derivatives. These are compounds containing a phenylphosphine, which consists of phosphine substituent bound to a phenyl group. . Phosphonates (Pn) are a large class of organophosphorus molecules that have direct carbon-phosphorus (C-P) bonds in place of the carbon-oxygen-phosphorus ester bond. In bacteria two pathways exist for Pn breakdown for use as a P source: the phosphonatase and C-P lyase pathways. These pathways differ both in regard to their substrate specificity and their cleavage mechanism. The phosphonatase pathway acts on the natural Pn alpha-aminoethylphosphonate (AEPn). In a two-step process it leads to cleavage of the C-P bond by a hydrolysis reaction requiring an adjacent carbonyl group. In contrast the C-P lyase pathway has a broad substrate specificity. It leads to cleavage of substituted Pn (such as AEPn) as well as unsubstituted Pn by a mechanism involving redox or radical chemistry. Due to its broad substrate specificity, the C-P lyase pathway is generally thought to be responsible for the breakdown of Pn herbicides (such as glyphosate) by bacteria.
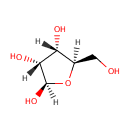
beta-D-ribofuranose (PAMDB001172)
IUPAC:
(2R,3R,4S,5R)-5-(hydroxymethyl)oxolane-2,3,4-triol
CAS: 50-69-1
Description: Beta-D-ribofuranose is the cyclic form of D-ribose. Ribose, primarily occurring as D-ribose, is an organic compound that occurs widely in nature. It is an aldopentose, a monosaccharide containing five carbon atoms that in its acyclic form has an aldehyde functional group at one end. Typically, ribose exists in the cyclic form. It comprises the backbone of RNA, a biopolymer that is the basis of genetic transcription. It is related to deoxyribose, as found in DNA, by the removal of one hydroxy group. Once phosphorylated, ribose can become a subunit of ATP, NADH, and several other compounds that are critical to metabolism.
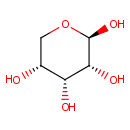
beta-D-Ribopyranose (PAMDB001173)
IUPAC:
(2R,3R,4R,5R)-oxane-2,3,4,5-tetrol
CAS: 50-69-1
Description: Beta-D-ribopyranose is a member of the chemical class known as Pentoses. These are monosaccharides in which the carbohydrate moiety contains five carbon atoms. Ribose 5-phosphate is involved in the pentose phosphate pathway and ribose degradation. The pentose phosphate pathway plays several key roles in metabolism including supply of biosynthetic carbon skeletons and reducing power. (PMID 9546650) Ribose-5-phosphate isomerase A (RpiA) plays an important role in interconverting between ribose-5-phosphate (R5P) and ribulose-5-phosphate in the pentose phosphate pathway and the Calvin cycle. (PMID 19214439). In Pseudomonas aeruginosa, RpiA catalyzes the interconversion of ribose-5-phosphate and ribulose-5-phosphate and is a key enzyme in the pentose phosphate pathway. (PMID 12182339).
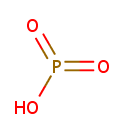
Phosphonate (PAMDB001286)
IUPAC:
oxophosphinic acid
CAS: 13598-36-2
Description: Phosphonates are one of the three sources of phosphate intake in biological cells (The other two being inorganic phosphate and organophosphate)
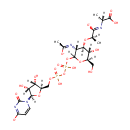
UDP-N-Acetylmuramyl-L-Ala (PAMDB001300)
IUPAC:
(2R)-N-[(1R)-1-carboxyethyl]-2-{[(3R,4R,5S,6R)-2-({[({[(2R,3S,4R,5R)-3,4-dihydroxy-5-(4-oxido-2-oxo-1,2-dihydropyrimidin-1-yl)oxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy](hydroxy)phosphoryl}oxy)-5-hydroxy-6-(hydroxymethyl)-3-[(1-oxidoethylidene)amino]oxan-4-yl]oxy}propanecarboximidate
CAS: Not Available
Description: UDP-n-acetylmuramyl-L-Ala is a precursor to peptidoglycan synthesis. UDP-N-acetylmuramoyl-L-alanine (UDP-MurNAc-L-Ala), which is the nucleotide substrate of the D-glutamic-acid-adding enzyme (the murD gene product) catalyzes key steps in the pathway for peptidoglycan synthesis. (PMID 8098327) MurD (UDP-N-acetylmuramoyl-L-alanine:D-glutamate ligase) is the second enzyme in the series of Mur ligases, and it catalyzes the addition of D-glutamic acid (D-Glu) to the cytoplasmic intermediate UDP-N-acetylmuramoyl-L-alanine (UMA). (PMID 19007109) The peptidoglycan synthesis pathway starts at the cytoplasm, where in six steps the peptidoglycan precursor a UDP-N-acetylmuramoyl-pentapeptide is synthesized. This precursor is then attached to the memberane acceptor all-trans-undecaprenyl phosphate, generating a N-acetylmuramoyl-pentapeptide-diphosphoundecaprenol, also known as lipid I. Another transferase then adds UDP-N-acetyl-D-glucosamine, yielding the complete monomeric unit a lipid II, also known as lipid II. This final lipid intermediate is transferred through the membrane. The peptidoglycan monomers are then polymerized on the outside surface by glycosyltransferases, which form the linear glycan chains, and transpeptidases, which catalyze the formation of peptide crosslinks.
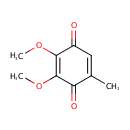
Ubiquinone-0 (PAMDB001302)
IUPAC:
2,3-dimethoxy-5-methylcyclohexa-2,5-diene-1,4-dione
CAS: 605-94-7
Description: Ubiquinone-1 is a member of the chemical class known as Benzoquinones. Normally in Pseudomonas aeruginosa the active form of Ubiquinone has 8 isoprene units (Ubiquinone-8) and in humans it normally has 10. Ubiquinone-10 is a naked version of Ubiquinone 8 that arises from conjugation by a shortened prenyl tail via 4-hydroxybenzoate polyprenyltransferase. Ubiquionone is involved in cellular respiration. It is fat-soluble and is therefore mobile in cellular membranes; it plays a unique role in the electron transport chain (ETC). In the inner bacterial membrane, electrons from NADH and succinate pass through the ETC to the oxygen, which is then reduced to water. The transfer of electrons through ETC results in the pumping of H+ across the membrane creating a proton gradient across the membrane, which is used by ATP synthase (located on the membrane) to generate ATP.
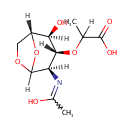
1,6-Anhydro-N-acetylmuramate (PAMDB001327)
IUPAC:
2-{[(1R,2S,3R,4R)-2-hydroxy-4-[(1-hydroxyethylidene)amino]-6,8-dioxabicyclo[3.2.1]octan-3-yl]oxy}propanoic acid
CAS: Not Available
Description: 1,6-anhMurNAC is an Amino Sugar. These are compounds having one alcoholic hydroxy group replaced by an amino group; systematically known as x-amino-x-deoxymonosaccharides. 1,6-anhydro-N-acetylmuramic acid (anhMurNAc) is one of the components of murein that is recycled inside the cell. Although exogenously provided anhMurNAc can be taken up by Pseudomonas aeruginosa, it can not serve as the sole source of carbon for growth.