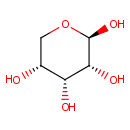
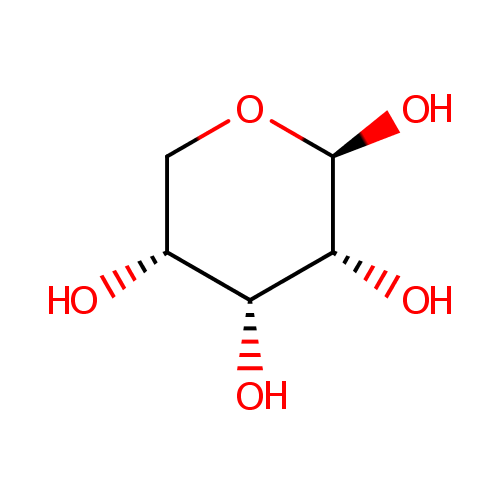
beta-D-Ribopyranose (PAMDB001173)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB001173 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | beta-D-Ribopyranose | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | Beta-D-ribopyranose is a member of the chemical class known as Pentoses. These are monosaccharides in which the carbohydrate moiety contains five carbon atoms. Ribose 5-phosphate is involved in the pentose phosphate pathway and ribose degradation. The pentose phosphate pathway plays several key roles in metabolism including supply of biosynthetic carbon skeletons and reducing power. (PMID 9546650) Ribose-5-phosphate isomerase A (RpiA) plays an important role in interconverting between ribose-5-phosphate (R5P) and ribulose-5-phosphate in the pentose phosphate pathway and the Calvin cycle. (PMID 19214439). In Pseudomonas aeruginosa, RpiA catalyzes the interconversion of ribose-5-phosphate and ribulose-5-phosphate and is a key enzyme in the pentose phosphate pathway. (PMID 12182339). | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C5H10O5 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 150.1299 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 150.05282343 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | SRBFZHDQGSBBOR-TXICZTDVSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C5H10O5/c6-2-1-10-5(9)4(8)3(2)7/h2-9H,1H2/t2-,3-,4-,5-/m1/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 50-69-1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | (2R,3R,4R,5R)-oxane-2,3,4,5-tetrol | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | β-D-ribopyranose | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | O[C@@H]1CO[C@@H](O)[C@H](O)[C@@H]1O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as monosaccharides. These are compounds containing one carbohydrate unit not glycosidically linked to another such unit, and no set of two or more glycosidically linked carbohydrate units. Monosaccharides have the general formula CnH2nOn. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organooxygen compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Carbohydrates and carbohydrate conjugates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Monosaccharides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Monosaccharides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic heteromonocyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | 0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | 95 °C | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in nucleotide binding
- Specific function:
- Part of the ABC transporter complex RbsABCD involved in ribose import. Responsible for energy coupling to the transport system
- Gene Name:
- rbsA
- Locus Tag:
- PA1947
- Molecular weight:
- 55.8 kDa
Reactions
| ATP + H(2)O + monosaccharide(Out) = ADP + phosphate + monosaccharide(In). |
- General function:
- Involved in transporter activity
- Specific function:
- Part of the binding-protein-dependent transport system for ribose. Probably responsible for the translocation of the substrate across the membrane
- Gene Name:
- rbsC
- Locus Tag:
- PA1948
- Molecular weight:
- 34 kDa
- General function:
- Carbohydrate transport and metabolism
- Specific function:
- Involved in the high-affinity D-ribose membrane transport system and also serves as the primary chemoreceptor for chemotaxis
- Gene Name:
- rbsB
- Locus Tag:
- PA1946
- Molecular weight:
- 33.9 kDa
Transporters
- General function:
- Involved in nucleotide binding
- Specific function:
- Part of the ABC transporter complex RbsABCD involved in ribose import. Responsible for energy coupling to the transport system
- Gene Name:
- rbsA
- Locus Tag:
- PA1947
- Molecular weight:
- 55.8 kDa
Reactions
| ATP + H(2)O + monosaccharide(Out) = ADP + phosphate + monosaccharide(In). |
- General function:
- Involved in transporter activity
- Specific function:
- Part of the binding-protein-dependent transport system for ribose. Probably responsible for the translocation of the substrate across the membrane
- Gene Name:
- rbsC
- Locus Tag:
- PA1948
- Molecular weight:
- 34 kDa
- General function:
- Carbohydrate transport and metabolism
- Specific function:
- Involved in the high-affinity D-ribose membrane transport system and also serves as the primary chemoreceptor for chemotaxis
- Gene Name:
- rbsB
- Locus Tag:
- PA1946
- Molecular weight:
- 33.9 kDa