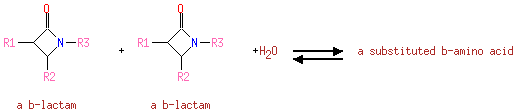| Identification |
| Name: |
1,6-anhydro-N-acetylmuramyl-L-alanine amidase AmpD |
| Synonyms: |
- N-acetylmuramoyl-L-alanine amidase
|
| Gene Name: |
ampD |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in N-acetylmuramoyl-L-alanine amidase activity |
| Specific Function: |
Involved in both cell wall peptidoglycans recycling and beta-lactamase induction. Specifically cleaves the amide bond between the lactyl group of N-acetylmuramic acid and the alpha- amino group of the L-alanine in degradation products containing an anhydro N-acetylmuramyl moiety |
| Cellular Location: |
Cytoplasm |
| KEGG Pathways: |
|
| KEGG Reactions: |
Not Available |
| SMPDB Reactions: |
|
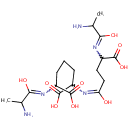
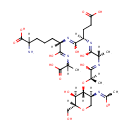
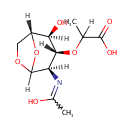
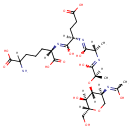
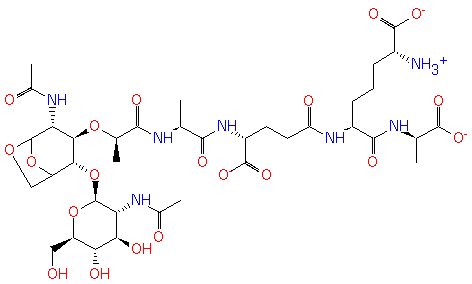
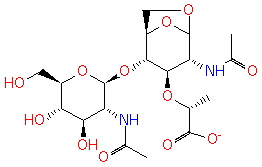
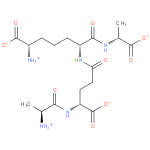
GlcNAc-1,6-anhMurNAc-L-Ala-?-D-Glu-DAP-D-Ala | + |  | → | N-acetyl-β-D-glucosamine(anhydrous)-N-acetylmuramate | + | L-Ala-gamma-D-Glu-DAP-D-Ala |
| GlcNAc-1,6-anhMurNAc-L-Ala-?-D-Glu-DAP-D-Ala + Water → N-acetyl-β-D-glucosamine(anhydrous)-N-acetylmuramate + L-Ala-gamma-D-Glu-DAP-D-Ala ReactionCard |
|
| PseudoCyc/BioCyc Reactions: |
| | | | | | | | | | | | | a β-lactam + a β-lactam + H 2O = a substituted β-amino acid ReactionCard | | |
|
| Complex Reactions: |
|
| Transports: |
Not Available |
| Metabolites: |
| PAMDB ID | Name | View |
|---|
| PAMDB001327 | 1,6-Anhydro-N-acetylmuramate | MetaboCard | | PAMDB001520 | 1,6-Anhydrous-N-Acetylmuramyl-tetrapeptide | MetaboCard | | PAMDB001521 | 1,6-Anhydrous-N-Acetylmuramyl-tripeptide | MetaboCard | | PAMDB001648 | L-alanine-D-glutamate-meso-2,6-diaminoheptanedioate | MetaboCard | | PAMDB001647 | L-Alanine-D-glutamate-meso-2,6-diaminoheptanedioate-D-alanine | MetaboCard | | PAMDB001656 | N-Acetyl-D-glucosamine(anhydrous)N-Acetylmuramic acid | MetaboCard | | PAMDB001657 | N-Acetyl-D-glucosamine(anhydrous)N-Acetylmuramyl-tetrapeptide | MetaboCard | | PAMDB001658 | N-Acetyl-D-glucosamine(anhydrous)N-Acetylmuramyl-tripeptide | MetaboCard | | PAMDB000142 | Water | MetaboCard |
|
| GO Classification: |
| Function |
|---|
| catalytic activity | | hydrolase activity | | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds | | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides | | N-acetylmuramoyl-L-alanine amidase activity | | Process |
|---|
| aminoglycan metabolic process | | carbohydrate metabolic process | | glycosaminoglycan metabolic process | | metabolic process | | peptidoglycan catabolic process | | peptidoglycan metabolic process | | polysaccharide metabolic process | | primary metabolic process |
|
| Gene Properties |
| Locus tag: |
PA4522 |
| Strand: |
- |
| Entrez Gene ID: |
881105 |
| Accession: |
NP_253212.1 |
| GI: |
15599718 |
| Sequence start: |
5064774 |
| Sequence End: |
5065340 |
| Sequence Length: |
566 |
| Gene Sequence: |
>PA4522
ATGCATTTCGATTCCGTTACCGGCTGGGTCCGTGGCGTCCGCCACTGCCCTTCACCGAATTTCAACCTGCGTCCGCAAGGGGACGCGGTGTCGCTGCTGGTTATCCACAATATCAGCCTGCCGCCGGGACAGTTCGGCACCGGTAAGGTCCAGGCGTTCTTCCAGAATCGCCTGGACCCGAACGAACATCCCTATTTCGAGGAGATCCGCCACCTGACGGTTTCCGCGCATTTCCTCATCGAACGCGACGGCGCCATCACCCAGTTCGTCTCCTGTCACGACCGCGCCTGGCATGCCGGGGTTTCCTGCTTCGACGGTCGCGAGGCGTGCAACGACTTCTCCCTTGGCATCGAACTGGAGGGGACCGACACGGAACCGTACACGGACGCGCAGTACACTGCCCTGGCCGGGCTGACGCGGCTGTTGCGCGCGGCCTTCCCGGGGATCACCCCGGAACGCATCCAGGGCCACTGCGATATCGCCCCGGAGCGCAAGACCGATCCCGGCGAGGCTTTCGACTGGTCGCGCTACCGCGCCGGATTGACCGATTCCAAGGAGGAAACATGA |
| Protein Properties |
| Protein Residues: |
188 |
| Protein Molecular Weight: |
21 kDa |
| Protein Theoretical pI: |
5.5 |
| Hydropathicity (GRAVY score): |
-0.447 |
| Charge at pH 7 (predicted): |
-6.99 |
| Protein Sequence: |
>PA4522
MHFDSVTGWVRGVRHCPSPNFNLRPQGDAVSLLVIHNISLPPGQFGTGKVQAFFQNRLDPNEHPYFEEIRHLTVSAHFLIERDGAITQFVSCHDRAWHAGVSCFDGREACNDFSLGIELEGTDTEPYTDAQYTALAGLTRLLRAAFPGITPERIQGHCDIAPERKTDPGEAFDWSRYRAGLTDSKEET |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA4522 and its homologs
|