| Identification |
| Name: |
Dihydroorotate dehydrogenase |
| Synonyms: |
- DHOdehase
- DHOD
- DHODase
- Dihydroorotate oxidase
|
| Gene Name: |
pyrD |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in catalytic activity |
| Specific Function: |
(S)-dihydroorotate + a quinone = orotate + a quinol |
| Cellular Location: |
Cell membrane; Peripheral membrane protein |
| KEGG Pathways: |
|
| KEGG Reactions: |
|
| SMPDB Reactions: |
|
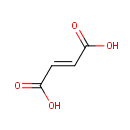
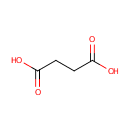
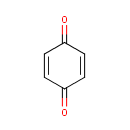
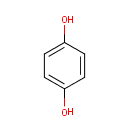
4,5-Dihydroorotic acid | + | 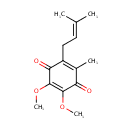 | + | 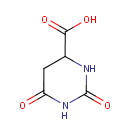 | → | 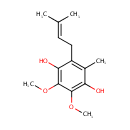 | + | 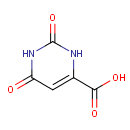 |
| |
|
| PseudoCyc/BioCyc Reactions: |
|
 | + | a ubiquinone | → |  | + | a ubiquinol |
| | |
 | + | a menaquinone | → |  | + | a menaquinol |
| |
|
| Complex Reactions: |
|
| Transports: |
Not Available |
| Metabolites: |
|
| GO Classification: |
| Component |
|---|
| cell part | | membrane | | Function |
|---|
| catalytic activity | | dihydroorotate dehydrogenase activity | | dihydroorotate oxidase activity | | oxidoreductase activity | | oxidoreductase activity, acting on the CH-CH group of donors | | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor | | Process |
|---|
| 'de novo' pyrimidine base biosynthetic process | | cellular aromatic compound metabolic process | | cellular metabolic process | | cellular nitrogen compound metabolic process | | metabolic process | | nitrogen compound metabolic process | | nucleobase metabolic process | | nucleobase, nucleoside and nucleotide metabolic process | | nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | | nucleoside phosphate metabolic process | | nucleotide metabolic process | | oxidation reduction | | pyrimidine base biosynthetic process | | pyrimidine base metabolic process | | pyrimidine nucleoside monophosphate biosynthetic process | | pyrimidine nucleotide biosynthetic process | | pyrimidine nucleotide metabolic process | | pyrimidine ribonucleoside monophosphate biosynthetic process | | UMP biosynthetic process |
|
| Gene Properties |
| Locus tag: |
PA3050 |
| Strand: |
- |
| Entrez Gene ID: |
882867 |
| Accession: |
NP_251740.1 |
| GI: |
15598246 |
| Sequence start: |
3414701 |
| Sequence End: |
3415729 |
| Sequence Length: |
1028 |
| Gene Sequence: |
>PA3050
ATGTACACCCTGGCCCGCCAGCTGCTGTTCAAACTGTCCCCGGAAACCTCCCACGAACTGTCCATCGACCTGATCGGGGCCGGCGGCCGCCTGGGCCTGAACCGCCTGCTGACGCCCAAGCCGGCCAGTCTGCCGGTCTCTGTGCTGGGCCTGGAGTTTCCCAACCCGGTGGGGCTGGCCGCGGGCCTGGACAAGAACGGCGACGCCATCGACGGGTTCGGCCAGCTCGGTTTCGGCTTCATCGAGATCGGCACCGTCACTCCGCGACCGCAGCCGGGCAATCCCAGGCCGCGCCTGTTCCGCCTGCCGCAAGCCAACGCGATCATCAATCGCATGGGTTTCAACAACCACGGCGTCGACCACCTGCTGGCGCGGGTGCGTGCGGCGAAGTATCGCGGCGTGCTGGGGATCAACATCGGCAAGAACTTCGATACCCCGGTGGAGCGCGCGGTCGACGACTACCTGATCTGCCTGGACAAGGTCTACGCCGACGCCAGCTACGTCACCGTCAACGTCAGCTCGCCGAACACGCCCGGCCTGCGCAGCCTGCAGTTCGGCGATTCGCTCAAGCAACTGCTGGAGGCACTGCGCCAGCGTCAGGAAGCGCTGGCGCTGCGGCATGGCCGGCGGGTACCGCTGGCGATCAAGATCGCCCCGGACATGACCGACGAAGAGACCGCGCTGGTCGCCGCGGCGCTGGTCGAGGCGGGGATGGACGCGGTGATCGCGACCAATACCACTCTCGGCCGCGAAGGCGTGGAAGGCCTGCCGCATGGCGACGAGGCGGGTGGCCTGTCCGGCGCGCCGGTGCGGGAAAAGAGCACGCACACGGTGAAGGTGCTGGCCGGCGAACTGGGCGGACGGCTGCCGATCATCGCCGCCGGCGGTATCACCGAGGGCGCGCACGCGGCGGAGAAGATCGCCGCCGGGGCGAGCCTGGTGCAGATCTATTCGGGTTTCATCTACAAGGGACCGGCGTTGATCCGCGAAGCGGTGGACGCCATCGCCGCATTGCCGCGGCGGAATTGA |
| Protein Properties |
| Protein Residues: |
342 |
| Protein Molecular Weight: |
36.1 kDa |
| Protein Theoretical pI: |
8.93 |
| Hydropathicity (GRAVY score): |
0.027 |
| Charge at pH 7 (predicted): |
3.65 |
| Protein Sequence: |
>PA3050
MYTLARQLLFKLSPETSHELSIDLIGAGGRLGLNRLLTPKPASLPVSVLGLEFPNPVGLAAGLDKNGDAIDGFGQLGFGFIEIGTVTPRPQPGNPRPRLFRLPQANAIINRMGFNNHGVDHLLARVRAAKYRGVLGINIGKNFDTPVERAVDDYLICLDKVYADASYVTVNVSSPNTPGLRSLQFGDSLKQLLEALRQRQEALALRHGRRVPLAIKIAPDMTDEETALVAAALVEAGMDAVIATNTTLGREGVEGLPHGDEAGGLSGAPVREKSTHTVKVLAGELGGRLPIIAAGGITEGAHAAEKIAAGASLVQIYSGFIYKGPALIREAVDAIAALPRRN |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA3050 and its homologs
|