| Identification |
| Name: |
uroporphyrin-III C-methyltransferase S-adenosyl-L-methionine:uroporphyrinogen III |
| Synonyms: |
Not Available |
| Gene Name: |
cobA |
| Enzyme Class: |
Not Available |
| Biological Properties |
| General Function: |
chlorophyll metabolic process, cofactor biosynthetic process, oxidation-reduction process, porphyrin-containing compound biosynthetic process, metabolic process |
| Specific Function: |
precorrin-2 dehydrogenase activity, methyltransferase activity |
| Cellular Location: |
Cytoplasmic |
| KEGG Pathways: |
- adenosylcobalamin biosynthesis II (late cobalt incorporation) P381-PWY
- cob(II)yrinate a,c-diamide biosynthesis II (late cobalt incorporation) PWY-7376
- cob(II)yrinate a,c-diamide biosynthesis I (early cobalt insertion) PWY-7377
- siroheme biosynthesis PWY-5194
|
| KEGG Reactions: |
Not Available |
| SMPDB Reactions: |
Not Available |
| PseudoCyc/BioCyc Reactions: |
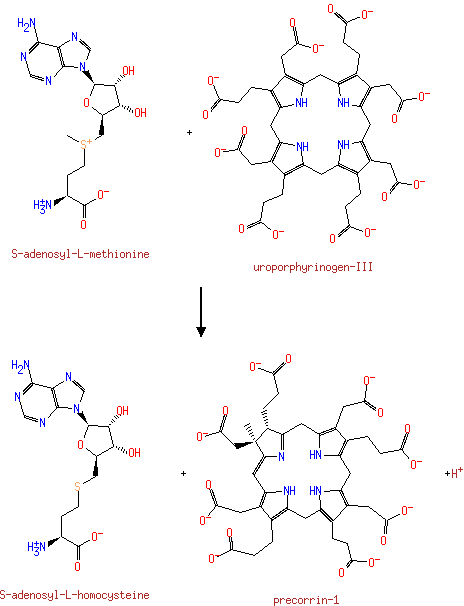
| S-adenosyl-L-methionine + uroporphyrinogen-III → S-adenosyl-L-homocysteine + precorrin-1 + H +ReactionCard | | |
|
| Complex Reactions: |
Not Available |
| Transports: |
Not Available |
| Metabolites: |
Not Available |
| GO Classification: |
| Function |
|---|
| precorrin-2 dehydrogenase activity | | methyltransferase activity | | Process |
|---|
| chlorophyll metabolic process | | cofactor biosynthetic process | | oxidation-reduction process | | porphyrin-containing compound biosynthetic process | | metabolic process |
|
| Gene Properties |
| Locus tag: |
PA1778 |
| Strand: |
- |
| Entrez Gene ID: |
877770 |
| Accession: |
NP_250469.1 |
| GI: |
15596975 |
| Sequence start: |
1922295 |
| Sequence End: |
1923032 |
| Sequence Length: |
737 |
| Gene Sequence: |
>PA1778
ATGAGCGGTAAGGTCTGGCTGGTGGGTGCGGGTCCCGGCGATCCGGAACTGTTGACCCTGAAGGCAGTTCGCGCCCTGCAGGACGCGGATGTGGTGATGGTCGACGATCTGGTCAACCCGAGCATACTGGAGCACTGTCCGTCCGCCCGGCTGGTCCGGGTCGGCAAACGCGGCGGCTGCCGCTCCACACCGCAGGACTTCATCCAGCGCCTGATGCTGCGGCATGCCCGCCAGGGACGCAGCGTGGTGCGCCTGAAAGGCGGCGATCCATGCATCTTCGGCCGCGCCGGCGAAGAAGCCGAATGGCTGGCCCGGCACGGGATAGACAGCGAGATAGTGAACGGCATCACCGCCGGGCTGGCTGGCGCCACTGCCTGCGGCATACCGCTGACCTACCGCGGCATCAGCCGCGGCGTGACCCTGGTGACCGCGCATACCCAGGACGACAGCCCGCTGGCCTGGGAAGCCCTGGCACGCAGCGGTACCACGCTGGTGGTCTACATGGGCGTCGCCCGTCTCGCAGAGATCCAGGCCGGGCTGCTGGCTGGAGGTATGGCCGAGGATACGCCGCTGGCGATGATCGAGAACGCCACCCTGGGCAACCAGCGCGAATGCCGCAGCAACCTCGGCGAGCTACTGCGGGACGCCGGACGTTTCGCCTTGAAAAGCCCGGCGATCCTGGTGATCGGCGAAGTAACCCGCGACATCGTGTCGCAACCGATTTCCCTGAGCGCCTGA |
| Protein Properties |
| Protein Residues: |
245 |
| Protein Molecular Weight: |
25.9 kDa |
| Protein Theoretical pI: |
7.41 |
| Hydropathicity (GRAVY score): |
0.091 |
| Charge at pH 7 (predicted): |
0.81 |
| Protein Sequence: |
>PA1778
MSGKVWLVGAGPGDPELLTLKAVRALQDADVVMVDDLVNPSILEHCPSARLVRVGKRGGCRSTPQDFIQRLMLRHARQGRSVVRLKGGDPCIFGRAGEEAEWLARHGIDSEIVNGITAGLAGATACGIPLTYRGISRGVTLVTAHTQDDSPLAWEALARSGTTLVVYMGVARLAEIQAGLLAGGMAEDTPLAMIENATLGNQRECRSNLGELLRDAGRFALKSPAILVIGEVTRDIVSQPISLSA |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA1778 and its homologs
|