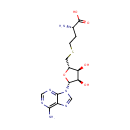
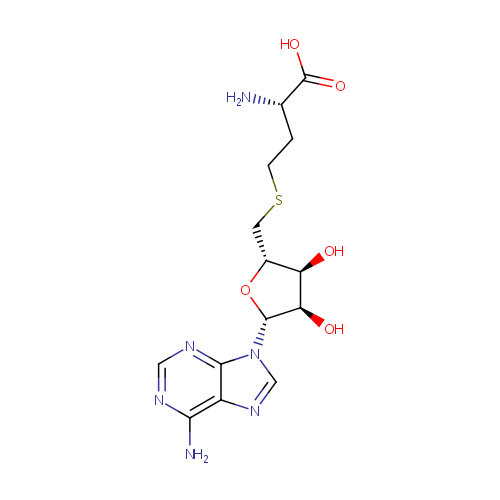
S-Adenosylhomocysteine (PAMDB000205)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000205 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | S-Adenosylhomocysteine | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | S-Adenosylhomocysteine (AdoHcy) is the immediate precursor of homocysteine. The reaction is catalyzed by S-adenosylhomocysteine hydrolase and is reversible with the equilibrium favoring formation of AdoHcy. In vivo, the reaction is driven in the direction of homocysteine formation by the action of the enzyme adenosine deaminase, which converts the second product of the S-adenosylhomocysteine hydrolase reaction, adenosine, to inosine. Except for methyl transfer from betaine and from methylcobalamin in the methionine synthase reaction, AdoHcy is the product of all methylation reactions that involve S-adenosylmethionine (AdoMet) as the methyl donor. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C14H20N6O5S | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 384.411 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 384.12158847 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | ZJUKTBDSGOFHSH-WFMPWKQPSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C14H20N6O5S/c15-6(14(23)24)1-2-26-3-7-9(21)10(22)13(25-7)20-5-19-8-11(16)17-4-18-12(8)20/h4-7,9-10,13,21-22H,1-3,15H2,(H,23,24)(H2,16,17,18)/t6-,7+,9+,10+,13+/m0/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 979-92-0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | (2S)-2-amino-4-({[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl}sulfanyl)butanoic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | S-adenosyl-L-homocysteine | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | N[C@@H](CCSC[C@H]1O[C@H]([C@H](O)[C@@H]1O)N1C=NC2=C(N)N=CN=C12)C(O)=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as glycosylamines. These are compounds consisting of an?amine?with a?beta-N-glycosidic bond?to a carbohydrate, thus forming a cyclic?hemiaminal ether?bond (alpha-amino ether). | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organooxygen compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Carbohydrates and carbohydrate conjugates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Glycosyl compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Glycosylamines | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic heteropolycyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | 0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | 209-211 °C | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | 2 S-Adenosylmethionine + Uroporphyrinogen III >2 S-Adenosylhomocysteine + Precorrin 2 + Hydrogen ion S-Adenosylhomocysteine + Water <> Adenine + S-Ribosyl-L-homocysteine S-Adenosylmethionine + L-Homocysteine + S-Methylmethionine <> S-Adenosylhomocysteine + Hydrogen ion + L-Methionine S-Adenosylmethionine + Malonyl-CoA > S-Adenosylhomocysteine + malonyl-CoA methyl ester trans-Aconitic acid + S-Adenosylmethionine > E-3-Carboxy-2-pentenedioate 6-methyl ester + S-Adenosylhomocysteine 2 S-Adenosylmethionine + PE(14:0/14:0) >2 S-Adenosylhomocysteine + Cyclopropane phosphatidylethanolamine (dihexadec-9,10-cyclo-anoyl, N-C16:0 cyclo) +2 Hydrogen ion 2 S-Adenosylmethionine + PE(14:0/14:0) >2 S-Adenosylhomocysteine + Cyclopropane phosphatidylethanolamine (dioctadec-11,12-cyclo-anoyl, N-C18:0 cyclo) +2 Hydrogen ion 2 S-Adenosylmethionine + PG(16:1(9Z)/16:1(9Z)) >2 S-Adenosylhomocysteine + Cyclopropane phosphatidylglycerol (dihexadec-9,10-cyclo-anoyl, N-C16:0 cyclo) +2 Hydrogen ion 2 S-Adenosylmethionine + PG(18:1(11Z)/18:1(11Z)) >2 S-Adenosylhomocysteine + Cyclopropane phosphatidylglycerol (dioctadec-11,12-cyclo-anoyl, N-C18:0 cyclo) +2 Hydrogen ion 2-Octaprenyl-6-hydroxyphenol + S-Adenosylmethionine > 2-Octaprenyl-6-methoxyphenol + S-Adenosylhomocysteine + Hydrogen ion 2-Octaprenyl-3-methyl-5-hydroxy-6-methoxy-1,4-benzoquinol + S-Adenosylmethionine > S-Adenosylhomocysteine + Hydrogen ion + Ubiquinol-8 2-Demethylmenaquinol 8 + S-Adenosylmethionine > S-Adenosylhomocysteine + Hydrogen ion + Menaquinol 8 2-Octaprenyl-6-methoxy-1,4-benzoquinol + S-Adenosylmethionine > 2-Octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol + S-Adenosylhomocysteine + Hydrogen ion S-Adenosylmethionine + L-Homocysteine <> S-Adenosylhomocysteine + L-Methionine 2 S-Adenosylmethionine + Uroporphyrinogen III <>2 S-Adenosylhomocysteine + Precorrin 2 S-Adenosylmethionine + DNA cytosine <> S-Adenosylhomocysteine + DNA 5-methylcytosine 2-Octaprenyl-6-hydroxyphenol + S-Adenosylmethionine <> 2-Octaprenyl-6-methoxyphenol + S-Adenosylhomocysteine 2-Demethylmenaquinone 8 + S-Adenosylmethionine <> Menaquinone + S-Adenosylhomocysteine 2-Phytyl-1,4-naphthoquinone + S-Adenosylmethionine <> Phylloquinone + S-Adenosylhomocysteine S-Adenosylmethionine + rRNA <> S-Adenosylhomocysteine + rRNA containing N6-methyladenine S-Adenosylmethionine + rRNA <> S-Adenosylhomocysteine + rRNA containing N1-methylguanine S-Adenosylmethionine + rRNA <> S-Adenosylhomocysteine + rRNA containing N2-methylguanine 2-Polyprenyl-6-hydroxyphenol + S-Adenosylmethionine <> 2-Polyprenyl-6-methoxyphenol + S-Adenosylhomocysteine More...2-Polyprenyl-6-methoxy-1,4-benzoquinone + S-Adenosylmethionine <> 2-Polyprenyl-3-methyl-6-methoxy-1,4-benzoquinone + S-Adenosylhomocysteine 2-Polyprenyl-3-methyl-5-hydroxy-6-methoxy-1,4-benzoquinone + S-Adenosylmethionine <> Ubiquinone-1 + S-Adenosylhomocysteine Demethylmenaquinol + S-Adenosylmethionine + Demethylmenaquinol <> Menaquinol 6 + S-Adenosylhomocysteine S-Adenosylmethionine + precorrin-1 > S-Adenosylhomocysteine + Precorrin 2 S-Adenosylmethionine + a [protein]-L-glutamine > Hydrogen ion + S-Adenosylhomocysteine + a [protein]-N<sup>5</sup>-methyl-L-glutamine tellurite + S-Adenosylmethionine methylated tellurite + S-Adenosylhomocysteine S-Adenosylmethionine + Uroporphyrinogen III <> S-Adenosylhomocysteine + precorrin-1 + Hydrogen ion S-Adenosylmethionine + a [protein]-L-β-isoaspartate > S-Adenosylhomocysteine + a protein L-β-isoaspartate α-methyl ester + Hydrogen ion a phospholipid olefinic fatty acid + S-Adenosylmethionine > a phospholipid cyclopropane fatty acid + S-Adenosylhomocysteine + Hydrogen ion L-Homocysteine + S-Adenosylmethionine Hydrogen ion + L-Methionine + S-Adenosylhomocysteine S-Adenosylmethionine + a demethylated methyl acceptor > S-Adenosylhomocysteine + a methylated methyl acceptor N-6-isopentyl adenosine-37 tRNA + S-Adenosylmethionine + <i>S</i>-sulfanyl-[acceptor] 2-methylthio-N-6-isopentyl adenosine-37 tRNA + S-Adenosylhomocysteine + L-Methionine + 5'-Deoxyadenosine + an unsulfurated sulfur acceptor + Hydrogen ion a guanine<sup>1516</sup> in 16S rRNA + S-Adenosylmethionine > an <i>N</i><sup>2</sup>-methylguanine<sup>1516</sup> in 16S rRNA + S-Adenosylhomocysteine + Hydrogen ion guanine<sup>2069</sup> in 23S rRNA + S-Adenosylmethionine > N<sup>7</sup>-methylguanine<sup>2069</sup> in 23S rRNA + S-Adenosylhomocysteine S-Adenosylmethionine <> S-Adenosylhomocysteine S-Adenosylmethionine + Phospholipid olefinic fatty acid <> S-Adenosylhomocysteine + Phospholipid cyclopropane fatty acid S-Adenosylmethionine + tRNA containing 5-aminomethyl-2-thiouridine <> S-Adenosylhomocysteine + tRNA containing 5-methylaminomethyl-2-thiouridylate S-Adenosylmethionine + DNA adenine <> S-Adenosylhomocysteine + DNA 6-methylaminopurine 2 S-Adenosylmethionine + Uroporphyrinogen III + Precorrin-1 <>2 S-Adenosylhomocysteine + Precorrin 2 2 S-Adenosylmethionine <> S-Adenosylhomocysteine +2 L-Methionine + 5'-Deoxyadenosine 2 S-Adenosylmethionine + Reduced acceptor <> S-Adenosylhomocysteine +2 L-Methionine + 5'-Deoxyadenosine S-Adenosylmethionine + Protein glutamate <> S-Adenosylhomocysteine + Protein glutamate methyl ester S-Adenosylmethionine <> S-Adenosylhomocysteine + DNA 5-methylcytosine S-Adenosylmethionine + Protein L-isoaspartate <> S-Adenosylhomocysteine + Protein L-isoaspartate methyl ester S-Adenosylmethionine + tRNA <> S-Adenosylhomocysteine + tRNA containing N6-methyladenine S-Adenosylhomocysteine + Water > Adenine + S-ribosyl-L-homocysteine + S-ribosyl-L-homocysteine a malonyl-[acp] + S-adenosyl-L-methionine > S-Adenosylhomocysteine + a malonyl-[acp] methyl ester Uroporphyrinogen III + S-adenosyl-L-methionine > Hydrogen ion + S-Adenosylhomocysteine + Precorrin-1 Precorrin-1 + S-adenosyl-L-methionine > Precorrin-2 + S-Adenosylhomocysteine 2-Octaprenyl-6-hydroxyphenol + S-adenosyl-L-methionine + 2-Octaprenyl-6-hydroxyphenol > Hydrogen ion + S-Adenosylhomocysteine + 2-methoxy-6-(all-trans-octaprenyl)phenol 3-demethylubiquinol-8 + S-adenosyl-L-methionine > Hydrogen ion + S-Adenosylhomocysteine + Ubiquinol 8 + Ubiquinol-8 2-Octaprenyl-6-methoxy-1,4-benzoquinol + S-adenosyl-L-methionine > S-Adenosylhomocysteine + Hydrogen ion + 6-Methoxy-3-methyl-2-all-trans-octaprenyl-1,4-benzoquinol 2-Demethylmenaquinol 8 + S-adenosyl-L-methionine > Hydrogen ion + S-Adenosylhomocysteine + Menaquinol 8 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Holy, Antonin; Rosenberg, Ivan. Studies on S-adenosyl-L-homocysteine hydrolase. Part XV. An improved synthesis of S-adenosyl-L-homocysteine and related compounds. Collection of Czechoslovak Chemical Communications (1985), 50(7), 1514-18. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Download (PDF) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||