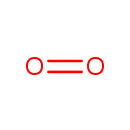| Identification |
| Name: |
Cytochrome o ubiquinol oxidase subunit 3 |
| Synonyms: |
- Cytochrome o ubiquinol oxidase subunit III
- Ubiquinol oxidase chain C
|
| Gene Name: |
cyoC |
| Enzyme Class: |
Not Available |
| Biological Properties |
| General Function: |
Involved in heme-copper terminal oxidase activity |
| Specific Function: |
Cytochrome o terminal oxidase complex is the component of the aerobic respiratory chain of E.coli that predominates when cells are grown at high aeration |
| Cellular Location: |
Cell inner membrane; Multi-pass membrane protein |
| KEGG Pathways: |
- Oxidative phosphorylation PW000919
- succinate to cytochrome bo oxidase electron transfer PWY0-1329
- proline to cytochrome bo oxidase electron transfer PWY0-1544
- NADH to cytochrome bo oxidase electron transfer PWY0-1335
|
| KEGG Reactions: |
Not Available |
| SMPDB Reactions: |
|
| PseudoCyc/BioCyc Reactions: |
| | | | | | |
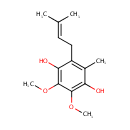
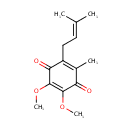
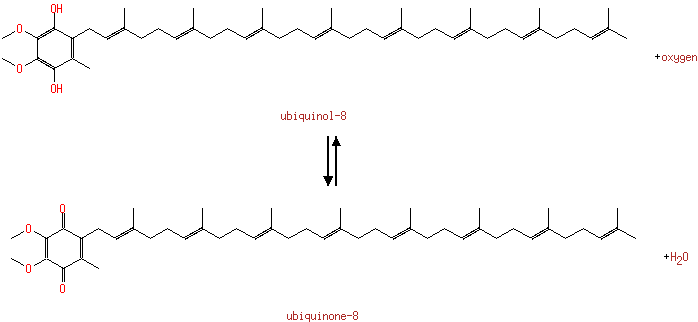
 | + | 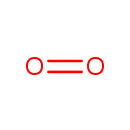 | + | Ubiquinols | → |  | + | Ubiquinones | + |  |
| |
|
| Complex Reactions: |
|
| Transports: |
Not Available |
| Metabolites: |
|
| GO Classification: |
| Component |
|---|
| cell part | | membrane | | plasma membrane | | Function |
|---|
| catalytic activity | | cytochrome-c oxidase activity | | heme-copper terminal oxidase activity | | oxidoreductase activity | | oxidoreductase activity, acting on diphenols and related substances as donors | | oxidoreductase activity, acting on diphenols and related substances as donors, oxygen as acceptor | | Process |
|---|
| cellular metabolic process | | electron transport chain | | generation of precursor metabolites and energy | | metabolic process | | mitochondrial electron transport, cytochrome c to oxygen | | oxidation reduction | | respiratory electron transport chain |
|
| Gene Properties |
| Locus tag: |
PA1319 |
| Strand: |
+ |
| Entrez Gene ID: |
881595 |
| Accession: |
NP_250010.1 |
| GI: |
15596516 |
| Sequence start: |
1431062 |
| Sequence End: |
1431691 |
| Sequence Length: |
629 |
| Gene Sequence: |
>PA1319
ATGTCGACGGCAGTATTGAACAAGCATCTGGCCGACGCCCACGAAGTGGGCCACGACCACGATCATGCACACGATAGCGGCGGCAACACCGTATTCGGTTTCTGGCTCTACCTGATGACCGACTGCGTGCTGTTCGCCAGCGTCTTCGCCACCTACGCGGTGCTGGTTCACCACACCGCCGGCGGCCCCAGCGGCAAGGACATCTTCGAGCTGCCCTACGTGCTGGTGGAAACCGCCATCCTGCTGGTTTCCAGCTGCACCTACGGCCTGGCCATGCTCTCTGCGCACAAGGGCGCCAAGGGCCAGGCGATCGCCTGGCTCGGGGTCACCTTCCTGCTCGGCGCCGCGTTCATCGGGATGGAGATCAACGAGTTCCACCACCTGATCGCCGAAGGTTTCGGCCCGAGCCGCAGCGCCTTCCTGTCGTCCTTCTTCACCCTGGTCGGCATGCACGGCCTGCACGTCAGCGCCGGTCTGCTGTGGATGCTGGTGCTGATGGCGCAGATCTGGACCCGCGGCCTCACCGCGCAGAACAACACCCGGATGATGTGCCTGAGCCTGTTCTGGCACTTCCTCGACATCGTCTGGATCTGCGTCTTCACCGTCGTCTACCTGATGGGGGCCCTGTAA |
| Protein Properties |
| Protein Residues: |
209 |
| Protein Molecular Weight: |
22.8 kDa |
| Protein Theoretical pI: |
6.62 |
| Hydropathicity (GRAVY score): |
0.695 |
| Charge at pH 7 (predicted): |
-2.77 |
| Protein Sequence: |
>PA1319
MSTAVLNKHLADAHEVGHDHDHAHDSGGNTVFGFWLYLMTDCVLFASVFATYAVLVHHTAGGPSGKDIFELPYVLVETAILLVSSCTYGLAMLSAHKGAKGQAIAWLGVTFLLGAAFIGMEINEFHHLIAEGFGPSRSAFLSSFFTLVGMHGLHVSAGLLWMLVLMAQIWTRGLTAQNNTRMMCLSLFWHFLDIVWICVFTVVYLMGAL |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA1319 and its homologs
|