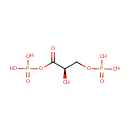| Identification |
| Name: |
Phosphoglycerate kinase |
| Synonyms: |
Not Available |
| Gene Name: |
pgk |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in phosphoglycerate kinase activity |
| Specific Function: |
ATP + 3-phospho-D-glycerate = ADP + 3-phospho- D-glyceroyl phosphate |
| Cellular Location: |
Cytoplasm |
| KEGG Pathways: |
- Carbon fixation in photosynthetic organisms pae00710
- Glycolysis / Gluconeogenesis pae00010
- Metabolic pathways pae01100
- Microbial metabolism in diverse environments pae01120
|
| KEGG Reactions: |
|
| SMPDB Reactions: |
|
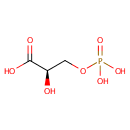
Glyceric acid 1,3-biphosphate | + | Adenosine diphosphate | + | 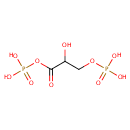 | + |  | → |  | + | 3-Phosphoglyceric acid | + | 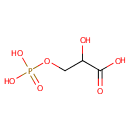 |
| | |
3-Phosphoglyceric acid | + |  | + |  | → | Adenosine diphosphate | + | Glyceric acid 1,3-biphosphate | + |  | + |  |
| | |
3-Phosphoglyceric acid | + |  | + |  | → | 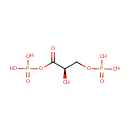 | + | Adenosine diphosphate | + |  |
| |
|
| PseudoCyc/BioCyc Reactions: |
|
| Complex Reactions: |
|
| Transports: |
Not Available |
| Metabolites: |
|
| GO Classification: |
| Function |
|---|
| catalytic activity | | kinase activity | | phosphoglycerate kinase activity | | transferase activity | | transferase activity, transferring phosphorus-containing groups | | Process |
|---|
| alcohol metabolic process | | glucose catabolic process | | glucose metabolic process | | glycolysis | | hexose metabolic process | | metabolic process | | monosaccharide metabolic process | | small molecule metabolic process |
|
| Gene Properties |
| Locus tag: |
PA0552 |
| Strand: |
+ |
| Entrez Gene ID: |
879198 |
| Accession: |
NP_249243.1 |
| GI: |
15595749 |
| Sequence start: |
611281 |
| Sequence End: |
612444 |
| Sequence Length: |
1163 |
| Gene Sequence: |
>PA0552
ATGACCGTGTTGAAGATGACTGACCTCGACCTCAAAGGTAAGCGCGTGCTGATCCGCGAGGACCTCAACGTGCCCGTGAAGGACGGCCAGGTGCAGAGCGATGCACGGATCAAGGCCGCCCTGCCGACCCTCAAGCTGGCCCTGGAGAAGGGCGCGGCGGTGATGGTCTGCTCGCACCTGGGCCGTCCGACCGAGGGCGAGTTCTCCGCCGAGAACAGCCTGAAGCCGGTGGCCGAGTACCTGTCCAAGGCCCTGGGCCGCGAAGTGCCGCTGCTGGCCGACTACCTCGACGGCGTCGAGGTCAAGGCGGGCGACCTGGTGCTGTTCGAGAACGTGCGCTTCAACAAGGGCGAGAAGAAGAACGCCGACGAGCTGGCGCAGAAGTACGCCGCGCTGTGCGACGTGTTCGTGATGGATGCCTTCGGTACCGCCCACCGCGCCGAGGGGTCGACCCACGGCGTGGCCAGGTTCGCCAAGGTCGCCGCCGCCGGCCCGCTGCTGGCCGCCGAACTGGACGCGCTGGGCAAGGCGCTGGGCAACCCGGCGCGGCCGATGGCGGCGATCGTCGCCGGCTCCAAGGTTTCCACCAAGCTCGACGTGCTGAACAGCCTGGCCGGCATCTGCGACCAACTGATCGTCGGCGGCGGCATCGCCAACACCTTCCTCGCCGCGGCCGGACACAAGGTCGGCAAGTCACTGTACGAGGCCGACCTGGTCGAGACCGCCAAGGCCATCGCCGCCAAGGTCAAGGTGCCGCTGCCGGTGGACGTGGTGGTGGCCAAGGAATTCGCCGAGAGCGCCGTGGCGACCGTCAAGGCCATCGCCGAGGTCGCCGACGACGACATGATCCTCGACATCGGCCCGCAGACCGCCGCGCAGTTCGCCGAGCTGCTGAAGACCTCGAAGACCATCCTGTGGAACGGTCCGGTCGGGGTGTTCGAGTTCGACCAGTTCGGCGAGGGCACCCGTACCCTGGCCAATGCCATCGCCGATAGCGCGGCGTTCTCCATCGCCGGCGGCGGCGATACCCTGGCGGCGATCGACAAGTACGGCATCGCCGAGCGGATCTCCTATATTTCCACCGGTGGCGGCGCGTTCCTCGAGTTCGTCGAGGGCAAGGTCCTGCCGGCCGTGGAAATCCTGGAGCAGCGCGCCAAGGGCTGA |
| Protein Properties |
| Protein Residues: |
387 |
| Protein Molecular Weight: |
40.4 kDa |
| Protein Theoretical pI: |
5.06 |
| Hydropathicity (GRAVY score): |
0.206 |
| Charge at pH 7 (predicted): |
-7.11 |
| Protein Sequence: |
>PA0552
MTVLKMTDLDLKGKRVLIREDLNVPVKDGQVQSDARIKAALPTLKLALEKGAAVMVCSHLGRPTEGEFSAENSLKPVAEYLSKALGREVPLLADYLDGVEVKAGDLVLFENVRFNKGEKKNADELAQKYAALCDVFVMDAFGTAHRAEGSTHGVARFAKVAAAGPLLAAELDALGKALGNPARPMAAIVAGSKVSTKLDVLNSLAGICDQLIVGGGIANTFLAAAGHKVGKSLYEADLVETAKAIAAKVKVPLPVDVVVAKEFAESAVATVKAIAEVADDDMILDIGPQTAAQFAELLKTSKTILWNGPVGVFEFDQFGEGTRTLANAIADSAAFSIAGGGDTLAAIDKYGIAERISYISTGGGAFLEFVEGKVLPAVEILEQRAKG |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA0552 and its homologs
|