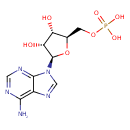
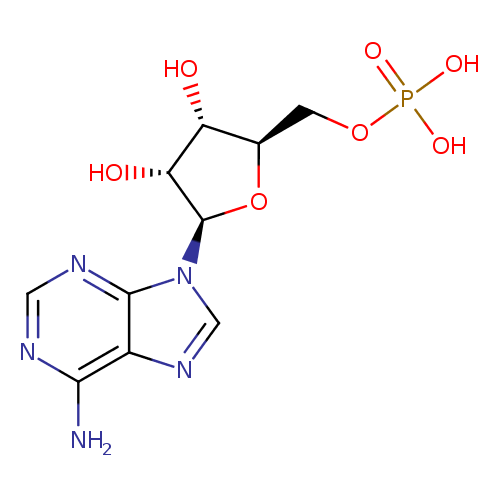
AMP (PAMDB110478)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB110478 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | AMP | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | A nucleoside 5'-monophosphate(2−) that results from the removal of two protons from the phosphate group of AMP. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C10H12N5O7P | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 345.21 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 347.0630843401 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | UDMBCSSLTHHNCD-KQYNXXCUSA-L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C10H14N5O7P/c11-8-5-9(13-2-12-8)15(3-14-5)10-7(17)6(16)4(22-10)1-21-23(18,19)20/h2-4,6-7,10,16-17H,1H2,(H2,11,12,13)(H2,18,19,20)/p-2/t4-,6-,7-,10-/m1/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 61-19-8 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | 5'-O-phosphonatoadenosine | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | adenylate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | C(C3(C(C(C(N2(C1(=C(C(=NC=N1)N)N=C2)))O3)O)O))OP([O-])([O-])=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of chemical entities known as purine ribonucleoside monophosphates. These are nucleotides consisting of a purine base linked to a ribose to which one monophosphate group is attached. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Chemical entities | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Nucleosides, nucleotides, and analogues | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Purine nucleotides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Purine ribonucleoside monophosphates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic heteropolycyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | 195 °C | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Odd-Straight-Chain-234-Sat-FA + ATP + coenzyme A → Odd-Saturated-Fatty-Acyl-CoA + AMP + diphosphate Citronellates + coenzyme A + ATP → citronellyll-CoA + AMP + diphosphate coenzyme A + acetoacetate + ATP → acetoacetyl-CoA + diphosphate + AMP thiamin + ATP → Hydrogen ion + thiamin diphosphate + AMP 3-Methyl-Saturated-Fatty-Acids + coenzyme A + ATP → 3-Methyl-Saturated-Fatty-Acyl-CoA + AMP + diphosphate ADP-D-ribose + Water → AMP + CPD-15317 + Hydrogen ion Water → AMP NADH + Water → Hydrogen ion + NMNH + AMP trans-cinnamate + coenzyme A + ATP → (E)-cinnamoyl-CoA + AMP + diphosphate tRNA-uridine34 + TusE-S-sulfanylcysteine + ATP → tRNA-2-thiouridine34 + TusE-L-cysteine + AMP + diphosphate Selenite + AMP + glutathione disulfide + Hydrogen ion → adenosine 5'-phosphoselenate + glutathione coenzyme A + propanoate + ATP → propanoyl-CoA + diphosphate + AMP linoleate + coenzyme A + ATP → linoleoyl-CoA + diphosphate + AMP L-Glutamine + L-aspartate + ATP + Water → Hydrogen ion + L-glutamate + L-Asparagine + diphosphate + AMP ATP + α-linolenate + coenzyme A → α-linolenoyl-CoA + diphosphate + AMP oleate + coenzyme A + ATP → oleoyl-CoA + AMP + diphosphate Long-Chain-Fatty-Acids + coenzyme A + ATP → Long-Chain-Acyl-CoAs + diphosphate + AMP undecaprenyl-diphospho-(N-acetylglucosamine)-N-acetylmuramoyl-L-alanyl-γ-D-isoglutaminyl-N-(β-D-asparatyl)-L-lysyl-D-alanyl-D-alanine + Ammonium + ATP → Hydrogen ion + undecaprenyl-diphospho-(N-acetylglucosamine)-N-acetylmuramoyl-L-alanyl-γ-D-isoglutaminyl-N-(β-D-asparaginyl)-L-lysyl-D-alanyl-D-alanine + AMP + diphosphate eicosapentaenoate + ATP + coenzyme A → eicosapentaenoyl-CoA + diphosphate + AMP 3-oxocholest-4-en-26-oate + ATP + coenzyme A → 3-oxocholest-4-en-26-oyl-CoA + AMP + diphosphate cyclic-AMP + Water → Hydrogen ion + AMP More...Hydrogen ion + L-Serine + ATP → diphosphate + AMP DEOXYNUCLEOTIDESM + ATP + Deoxynucleotides → AMP + diphosphate + Deoxynucleotides CPD66-39 + coenzyme A + ATP → Saturated-Fatty-Acyl-CoA + diphosphate + AMP phytenate + ATP + coenzyme A → phytenoyl-CoA + AMP + diphosphate ATP → AMP + diphosphate biotin + ATP → diphosphate + AMP phenylacetate + ATP + coenzyme A → phenylacetyl-CoA + AMP + diphosphate (R)-4'-phosphopantothenate + L-Cysteine + ATP → Hydrogen ion + R-4'-phosphopantothenoyl-L-cysteine + diphosphate + AMP R2OH-Straight-Chain-234-Sat-FA + ATP + coenzyme A → R2-2OH-Straight-Chain-234-Sat-FA-CoA + AMP + diphosphate (15Z)-tetracosenoate + coenzyme A + ATP → (Z)-15-tetracosenoyl-CoA + diphosphate + AMP O-ureidohomoserine + L-aspartate + ATP → Hydrogen ion + canavaninosuccinate + AMP + diphosphate palmitate + coenzyme A + ATP → palmitoyl-CoA + diphosphate + AMP GDP + ADP → GTP + AMP 3-[(3aS,4S,7aS)-7a-methyl-1,5-dioxo-octahydro-1H-inden-4-yl]propanoate + ATP + coenzyme A → 3-[(3aS,4S,7aS)-7a-methyl-1,5-dioxo-octahydro-1H-inden-4-yl]propanoyl-CoA + AMP + diphosphate L-glutamate + ATP + Hydrogen ion → AMP + diphosphate pristanate + coenzyme A + ATP → pristanoyl-CoA + diphosphate + AMP 2-Me-Branched-234-Sat-FA + coenzyme A + ATP → 2-Me-Branched-234-Sat-Fatty-Acyl-CoA + diphosphate + AMP β-D-fructofuranose 6-phosphate + ADP → Hydrogen ion + fructose 1,6-bisphosphate + AMP tRNAs-Asp-with-queuosine + ATP + L-glutamate → tRNAs-with-glutamylated-queuosine + AMP + diphosphate + Hydrogen ion Sulfite + AMP + Acceptor + Hydrogen ion → adenosine 5'-phosphosulfate + Donor-H2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Download (PDF) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||