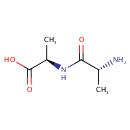
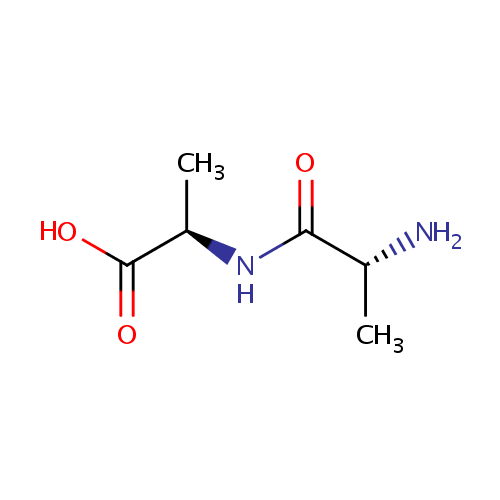
D-Alanyl-D-alanine (PAMDB000504)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000504 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | D-Alanyl-D-alanine | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | D-Alanyl-D-alanine is a component of peptidoglycan. The ATP-dependent carboxylate-amine/thiol ligase superfamily is known to contain enzymes catalyzing the formation of various types of peptide, one of which is D-alanyl-D-alanine. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C6H12N2O3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 160.1711 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 160.08479226 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | DEFJQIDDEAULHB-QWWZWVQMSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C6H12N2O3/c1-3(7)5(9)8-4(2)6(10)11/h3-4H,7H2,1-2H3,(H,8,9)(H,10,11)/t3-,4-/m1/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 923-16-0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | (2R)-2-[(2R)-2-aminopropanamido]propanoic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | D-ala-D-ala | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | C[C@@H](N)C(=O)N[C@H](C)C(O)=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as peptides. These are compounds containing an amide derived from two or more amino carboxylic acid molecules (the same or different) by formation of a covalent bond from the carbonyl carbon of one to the nitrogen atom of another. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Carboxylic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Amino acids, peptides, and analogues | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Peptides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | 0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Adenosine triphosphate + Water + D-Alanyl-D-alanine > ADP + D-Alanyl-D-alanine + Hydrogen ion + Phosphate Adenosine triphosphate + Water + D-Alanyl-D-alanine > ADP + D-Alanyl-D-alanine + Hydrogen ion + Phosphate two linked disacharide pentapeptide murein units (uncrosslinked, middle of chain) > D-Alanyl-D-alanine + two disacharide linked murein units, pentapeptide corsslinked tripeptide (A2pm->A2pm) (middle of chain) 2 D-Alanine + Adenosine triphosphate <> ADP + D-Alanyl-D-alanine + Hydrogen ion + Phosphate D-Alanyl-D-alanine + Adenosine triphosphate + UDP-N-Acetylmuramoyl-L-alanyl-D-glutamyl-meso-2,6-diaminoheptanedioate > ADP + Hydrogen ion + Phosphate + UDP-N-Acetylmuramoyl-L-alanyl-D-glutamyl-6-carboxy-L-lysyl-D-alanyl-D-alanine D-Alanyl-D-alanine + Water >2 D-Alanine Adenosine triphosphate + 2 D-Alanine <> ADP + Phosphate + D-Alanyl-D-alanine Adenosine triphosphate + UDP-N-Acetylmuramoyl-L-alanyl-gamma-D-glutamyl-L-lysine + D-Alanyl-D-alanine <> ADP + Phosphate + UDP-N-Acetylmuramoyl-L-alanyl-D-glutamyl-L-lysyl-D-alanyl-D-alanine Adenosine triphosphate + UDP-N-Acetylmuramoyl-L-alanyl-D-glutamyl-meso-2,6-diaminoheptanedioate + D-Alanyl-D-alanine <> ADP + Phosphate + UDP-N-Acetylmuramoyl-L-alanyl-D-glutamyl-6-carboxy-L-lysyl-D-alanyl-D-alanine Adenosine triphosphate + 2 D-Alanine > ADP + Inorganic phosphate + D-Alanyl-D-alanine Adenosine triphosphate + UDP-N-Acetylmuramoyl-L-alanyl-gamma-D-glutamyl-L-lysine + D-Alanyl-D-alanine > ADP + Inorganic phosphate + UDP-N-Acetylmuramoyl-L-alanyl-D-glutamyl-L-lysyl-D-alanyl-D-alanine 2 D-Alanine + Adenosine triphosphate > D-Alanyl-D-alanine + Adenosine diphosphate + Phosphate + ADP UDP-N-Acetylmuramoyl-L-alanyl-D-gamma-glutamyl-meso-2,6-diaminopimelate + D-Alanyl-D-alanine + Adenosine triphosphate > Adenosine diphosphate + Phosphate + Hydrogen ion + UDP-N-acetyl-α-D-muramoyl-L-alanyl-γ-D-glutamyl-meso-2,6-diaminopimeloyl-D-alanyl-D-alanine + ADP | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Neuhaus, Francis C. Enzymic synthesis of D-alanyl-D-alanine. Biochemical and Biophysical Research Communications (1960), 3 401-5. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Download (PDF) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in catalytic activity
- Specific function:
- Cell wall formation. Synthesis of cross-linked peptidoglycan from the lipid intermediates. The enzyme has a penicillin-insensitive transglycosylase N-terminal domain (formation of linear glycan strands) and a penicillin-sensitive transpeptidase C-terminal domain (cross-linking of the peptide subunits)
- Gene Name:
- mrcB
- Locus Tag:
- PA4700
- Molecular weight:
- 85.5 kDa
Reactions
| (GlcNAc-(1->4)-Mur2Ac(oyl-L-Ala-gamma-D-Glu-L-Lys-D-Ala-D-Ala))(n)-diphosphoundecaprenol + GlcNAc-(1->4)-Mur2Ac(oyl-L-Ala-gamma-D-Glu-L-Lys-D-Ala-D-Ala)-diphosphoundecaprenol = (GlcNAc-(1->4)-Mur2Ac(oyl-L-Ala-gamma-D-Glu-L-Lys-D-Ala-D-Ala))(n+1)-diphosphoundecaprenol + undecaprenyl diphosphate. |
- General function:
- Involved in ATP binding
- Specific function:
- Cell wall formation
- Gene Name:
- ddlB
- Locus Tag:
- PA4410
- Molecular weight:
- 34.4 kDa
Reactions
| ATP + 2 D-alanine = ADP + phosphate + D-alanyl-D-alanine. |
- General function:
- Involved in ATP binding
- Specific function:
- Cell wall formation
- Gene Name:
- ddlA
- Locus Tag:
- PA4201
- Molecular weight:
- 36.5 kDa
Reactions
| ATP + 2 D-alanine = ADP + phosphate + D-alanyl-D-alanine. |
- General function:
- Involved in ATP binding
- Specific function:
- Involved in cell wall formation. Catalyzes the final step in the synthesis of UDP-N-acetylmuramoyl-pentapeptide, the precursor of murein
- Gene Name:
- murF
- Locus Tag:
- PA4416
- Molecular weight:
- 47.4 kDa
Reactions
| ATP + UDP-N-acetylmuramoyl-L-alanyl-gamma-D-glutamyl-L-lysine + D-alanyl-D-alanine = ADP + phosphate + UDP-N-acetylmuramoyl-L-alanyl-gamma-D-glutamyl-L-lysyl-D-alanyl-D-alanine. |
- General function:
- Involved in transporter activity
- Specific function:
- Part of the binding-protein-dependent transport system for dipeptides; probably responsible for the translocation of the substrate across the membrane
- Gene Name:
- dppB
- Locus Tag:
- PA4503
- Molecular weight:
- 37 kDa
- General function:
- Involved in transporter activity
- Specific function:
- Part of the binding-protein-dependent transport system for dipeptides; probably responsible for the translocation of the substrate across the membrane
- Gene Name:
- dppC
- Locus Tag:
- PA4504
- Molecular weight:
- 32.4 kDa
- General function:
- Involved in nucleotide binding
- Specific function:
- Part of the binding-protein-dependent transport system for dipeptides. Probably responsible for energy coupling to the transport system
- Gene Name:
- dppD
- Locus Tag:
- PA4505
- Molecular weight:
- 35.4 kDa
- General function:
- Involved in nucleotide binding
- Specific function:
- Part of the binding-protein-dependent transport system for dipeptides. Probably responsible for energy coupling to the transport system
- Gene Name:
- dppF
- Locus Tag:
- PA4506
- Molecular weight:
- 36.3 kDa
- General function:
- Involved in transporter activity
- Specific function:
- Dipeptide-binding protein of a transport system that can be subject to osmotic shock. DppA is also required for peptide chemotaxis
- Gene Name:
- dppA
- Locus Tag:
- PA4496
- Molecular weight:
- 60.1 kDa
Transporters
- General function:
- Involved in transporter activity
- Specific function:
- Part of the binding-protein-dependent transport system for dipeptides; probably responsible for the translocation of the substrate across the membrane
- Gene Name:
- dppB
- Locus Tag:
- PA4503
- Molecular weight:
- 37 kDa
- General function:
- Involved in transporter activity
- Specific function:
- Part of the binding-protein-dependent transport system for dipeptides; probably responsible for the translocation of the substrate across the membrane
- Gene Name:
- dppC
- Locus Tag:
- PA4504
- Molecular weight:
- 32.4 kDa
- General function:
- Involved in nucleotide binding
- Specific function:
- Part of the binding-protein-dependent transport system for dipeptides. Probably responsible for energy coupling to the transport system
- Gene Name:
- dppD
- Locus Tag:
- PA4505
- Molecular weight:
- 35.4 kDa
- General function:
- Involved in nucleotide binding
- Specific function:
- Part of the binding-protein-dependent transport system for dipeptides. Probably responsible for energy coupling to the transport system
- Gene Name:
- dppF
- Locus Tag:
- PA4506
- Molecular weight:
- 36.3 kDa
- General function:
- Involved in transporter activity
- Specific function:
- Dipeptide-binding protein of a transport system that can be subject to osmotic shock. DppA is also required for peptide chemotaxis
- Gene Name:
- dppA
- Locus Tag:
- PA4496
- Molecular weight:
- 60.1 kDa