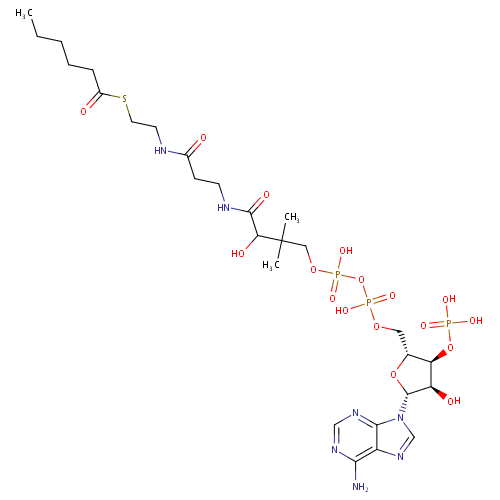
Hexanoyl-CoA (PAMDB000467)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000467 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Hexanoyl-CoA | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | Hexanoyl-CoA is a fatty acid coenzyme A derivative that can be involved in the biosynthesis and oxidation of fatty acids. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C27H46N7O17P3S | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 865.677 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 865.188373307 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | OEXFMSFODMQEPE-UHFFFAOYSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C27H46N7O17P3S/c1-4-5-6-7-18(36)55-11-10-29-17(35)8-9-30-25(39)22(38)27(2,3)13-48-54(45,46)51-53(43,44)47-12-16-21(50-52(40,41)42)20(37)26(49-16)34-15-33-19-23(28)31-14-32-24(19)34/h14-16,20-22,26,37-38H,4-13H2,1-3H3,(H,29,35)(H,30,39)(H,43,44)(H,45,46)(H2,28,31,32)(H2,40,41,42) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 5060-32-2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | {[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-2-[({[({3-[(2-{[2-(hexanoylsulfanyl)ethyl]carbamoyl}ethyl)carbamoyl]-3-hydroxy-2,2-dimethylpropoxy}(hydroxy)phosphoryl)oxy](hydroxy)phosphoryl}oxy)methyl]-4-hydroxyoxolan-3-yl]oxy}phosphonic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-2-{[({3-[(2-{[2-(hexanoylsulfanyl)ethyl]carbamoyl}ethyl)carbamoyl]-3-hydroxy-2,2-dimethylpropoxy(hydroxy)phosphoryl}oxy(hydroxy)phosphoryl)oxy]methyl}-4-hydroxyoxolan-3-yl]oxyphosphonic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | CCCCCC(=O)SCCN=C(O)CCN=C(O)C(O)C(C)(C)COP(O)(=O)OP(O)(=O)OCC1OC(C(O)C1OP(O)(O)=O)N1C=NC2=C(N)N=CN=C12 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as 2,3,4-saturated fatty acyl coas. These are acyl-CoAs carrying a 2,3,4-saturated fatty acyl chain. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Lipids and lipid-like molecules | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Fatty Acyls | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Fatty acyl thioesters | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | 2,3,4-saturated fatty acyl CoAs | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic heteropolycyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -4 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Adenosine triphosphate + Coenzyme A + Hydrogen ion + Hexanoate (N-C6:0) > Adenosine monophosphate + Hydrogen ion + Hexanoyl-CoA + Pyrophosphate Acetyl-CoA + Hexanoate (N-C6:0) > Acetic acid + Hexanoyl-CoA Acetyl-CoA + Hexanoyl-CoA <> 3-Oxooctanoyl-CoA + Coenzyme A FAD + Hexanoyl-CoA <> FADH2 + trans-2-Hexenoyl-CoA Water + Hexanoyl-CoA > Coenzyme A + Hydrogen ion + Hexanoate (N-C6:0) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Pullman, Maynard E. Convenient and versatile method for the purification of CoA thiol esters. Analytical Biochemistry (1973), 54(1), 188-98. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in acetyl-CoA C-acyltransferase activity
- Specific function:
- Catalyzes the final step of fatty acid oxidation in which acetyl-CoA is released and the CoA ester of a fatty acid two carbons shorter is formed. Involved in the aerobic and anaerobic degradation of long-chain fatty acids
- Gene Name:
- fadA
- Locus Tag:
- PA3013
- Molecular weight:
- 41.6 kDa
Reactions
| Acyl-CoA + acetyl-CoA = CoA + 3-oxoacyl-CoA. |
- General function:
- Involved in catalytic activity
- Specific function:
- Catalyzes the esterification, concomitant with transport, of exogenous long-chain fatty acids into metabolically active CoA thioesters for subsequent degradation or incorporation into phospholipids
- Gene Name:
- fadD
- Locus Tag:
- PA3299
- Molecular weight:
- 61.7 kDa
Reactions
| ATP + a long-chain fatty acid + CoA = AMP + diphosphate + an acyl-CoA. |
- General function:
- Involved in acyl-CoA dehydrogenase activity
- Specific function:
- Catalyzes the dehydrogenation of acyl-CoA
- Gene Name:
- fadE
- Locus Tag:
- PA2815
- Molecular weight:
- 88.8 kDa
Reactions
| An acyl-CoA + FAD = a dehydrogenated acyl-CoA + FADH(2). |
- General function:
- Involved in acyl-CoA hydrolase activity
- Specific function:
- Can hydrolyze a broad range of acyl-CoA thioesters. Its physiological function is not known
- Gene Name:
- tesB
- Locus Tag:
- PA3942
- Molecular weight:
- 32.9 kDa