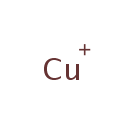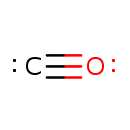
Search Results for compounds
Searching compounds for
returned 4373 results.
Displaying compounds 1791 - 1800 of
4373 in total
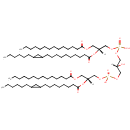
CL(14:0/19:0cycv8c/14:0/17:0cycw7c) (PAMDB003390)
IUPAC:
[(2R)-2-{[10-(2-hexylcyclopropyl)decanoyl]oxy}-3-(tetradecanoyloxy)propoxy][(2S)-3-({[(2R)-2-{[8-(2-hexylcyclopropyl)octanoyl]oxy}-3-(tetradecanoyloxy)propoxy](hydroxy)phosphoryl}oxy)-2-hydroxypropoxy]phosphinic acid
CAS: Not Available
Description: CL(14:0/19:0cycv8c/14:0/17:0cycw7c) is a cardiolipin (CL). Cardiolipins are sometimes called a 'double' phospholipid because they have four fatty acid tails, instead of the usual two. CL(14:0/19:0cycv8c/14:0/17:0cycw7c) contains two chains of tetradecanoic acid at the C1 and C3 positions, one chain of (heptadec-11-12-cyclo-anoyl) at the C2 position, one chain of (heptadec-9-10-cyclo-anoyl) at the C4 position. While the theoretical charge of cardiolipins is -2, under normal physiological conditions (pH near 7), the molecule may carry only one negative charge. In prokaryotes such as Pseudomonas aeruginosa, the enzyme known as diphosphatidylglycerol synthase catalyses the transfer of the phosphatidyl moiety of one phosphatidylglycerol to the free 3'-hydroxyl group of another, with the elimination of one molecule of glycerol. In Pseudomonas aeruginosa, which acylates its glycerophospholipids with acyl chains ranging in length from 12 to 19 carbons and possibly containing an unsaturation, or a cyclopropane group more than 100 possible CL molecular species are theoretically possible. Pseudomonas aeruginosa membranes consist of ~5% cardiolipin (CL), 20-25% phosphatidylglycerol (PG), and 70-80% phosphatidylethanolamine (PE) as well as smaller amounts of phosphatidylserine (PS). CL is distributed between the two leaflets of the bilayers and is located preferentially at the poles and septa in Pseudomonas aeruginosa and other rod-shaped bacteria. It is known that the polar positioning of the proline transporter ProP and the mechanosensitive ion channel MscS in Pseudomonas aeruginosa is dependent on CL. It is believed that cell shape may influence the localization of CL and the localization of certain membrane proteins.
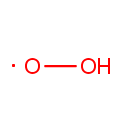
superoxide (PAMDB003391)
IUPAC:
dioxidanyl
CAS: 11062-77-4
Description: Highly reactive compounds produced when oxygen is reduced by a single electron. In biological systems, they may be generated during the normal catalytic function of a number of enzymes and during the oxidation of hemoglobin to METHEMOGLOBIN. In living organisms, SUPEROXIDE DISMUTASE protects the cell from the deleterious effects of superoxides.
Cu(+) (PAMDB003393)
IUPAC:
??-copper(1+) ion
CAS: 7440-50-8
Description: The copper (I) ion is typically found in Zn-Cu superoxide dismutase. In the SOD-catalysed dismutation of superoxide the oxidation state of copper oscillates between 1 and 2. Most soluble copper ions are Cu2+ and most Cu+ ions will disproportionate to Cu2+ ions. The disproportionation reaction only occurs with simple copper(I) ions in solution.
CO (PAMDB003394)
IUPAC:
Not Available
CAS: 124-38-9
Description: Carbon monoxide, with the chemical formula CO, is a colorless, odorless, and tasteless gas. It consists of one carbon atom covalently bonded to one oxygen atom. It is a gas at room temperature. Carbon monoxide is an intermediate in 4-amino-2-methyl-5-diphosphomethylpyrimidine biosynthesis in Pseudomonas aeruginosa. It is a product for enzyme HMP-P synthase which catalyzes the chemical reaction 5-amino-1-(5-phospho-beta-D-ribosyl)imidazole + S-adenosyl-L-methionine -> 4-amino-2-methyl-5-phosphomethylpyrimidine + 5'-deoxyadenosine + L-methionine + formate + carbon monoxide + 3 H+ (EcoCyc compound: CARBON-MONOXIDE).
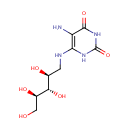
5-amino-6-(D-ribitylamino)uracil (PAMDB003406)
IUPAC:
5-amino-6-{[(2S,3S,4R)-2,3,4,5-tetrahydroxypentyl]amino}-1,2,3,4-tetrahydropyrimidine-2,4-dione
CAS: Not Available
Description: 5-amino-6-(D-ribitylamino)uracil is involved in the Cofactor riboflavin biosynthesis pathway. It is a substrate of Riboflavin biosynthesis protein RibD that catalyses the following reactions: 2-hydroxy-3-oxobutyl phosphate + 5-amino-6-(D-ribitylamino)uracil => 6,7-dimethyl-8-(1-D-ribityl)lumazine + H(2)O + 1 phosphate and 6,7-dimethyl-8-(1-D-ribityl)lumazine => 5-amino-6-(D-ribitylamino)uracil + riboflavin
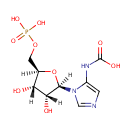
5-carboxyamino-1-(5-phospho-D-ribosyl)imidazole (PAMDB003410)
IUPAC:
{1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-[(phosphonooxy)methyl]oxolan-2-yl]-1H-imidazol-5-yl}carbamic acid
CAS: Not Available
Description: A 1-(phosphoribosyl)imidazole having the phospho group at the 5'-position and a carboxyamino group at the 5-position on the imidazole ring
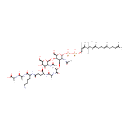
lipid II(A) (PAMDB003413)
IUPAC:
(4R)-N-[(1S,5R)-5-amino-5-carboxy-1-{[(1R)-1-{[(1R)-1-carboxyethyl]carboximidato}ethyl]-C-hydroxycarbonimidoyl}pentyl]-4-carboxy-4-{[(2S)-2-{[(2R)-2-{[(2R,3S,4R,5R,6R)-3-{[(2R,3R,4R,5S,6R)-4,5-dihydroxy-6-(hydroxymethyl)-3-[(1-oxidoethylidene)amino]oxan-2-yl]oxy}-6-{[hydroxy({[hydroxy({[(2Z,6Z,10Z,14Z,18Z,22Z,26Z,30Z,34E,38E)-3,7,11,15,19,23,27,31,35,39,43-undecamethyltetratetraconta-2,6,10,14,18,22,26,30,34,38,42-undecaen-1-yl]oxy})phosphoryl]oxy})phosphoryl]oxy}-2-(hydroxymethyl)-5-[(1-oxidoethylidene)amino]oxan-4-yl]oxy}-1-hydroxypropylidene]amino}-1-hydroxypropylidene]amino}butanecarboximidate
CAS: Not Available
Description: Lipid II is a membrane-anchored cell-wall precursor that is essential for bacterial cell-wall biosynthesis. It is the target for at least four different classes of antibiotic, including the clinically important glycopeptide antibiotic vancomycin. Lipid II consists of one GlcNAc-MurNAc-pentapeptide subunit linked to a polyiosoprenoid anchor 11 subunits long via a pyrophosphate linker.
GlcNAc-(1->4)-Mur2Ac(oyl-L-Ala-gamma-D-Glu-L-Lys-D-Ala-D-Ala)-diphosphoundecaprenol (PAMDB003414)
IUPAC:
(2R)-4-{[(1S)-5-amino-1-{[(1R)-1-{[(1R)-1-carboxyethyl]carbamoyl}ethyl]carbamoyl}pentyl]carbamoyl}-2-[(2S)-2-[(2R)-2-{[(2R,3R,4R,5S,6R)-3-acetamido-5-{[(2S,3R,4R,5S,6R)-3-acetamido-4,5-dihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}-2-{[hydroxy({[hydroxy({[(2Z,6Z,10Z,14Z,18Z,22Z,26Z,30Z,34E,38E)-3,7,11,15,19,23,27,31,35,39,43-undecamethyltetratetraconta-2,6,10,14,18,22,26,30,34,38,42-undecaen-1-yl]oxy})phosphoryl]oxy})phosphoryl]oxy}-6-(hydroxymethyl)oxan-4-yl]oxy}propanamido]propanamido]butanoic acid
CAS: Not Available
Description: GlcNAc-(1->4)-Mur2Ac(oyl-L-Ala-gamma-D-Glu-L-Lys-D-Ala-D-Ala)-diphosphoundecaprenol is an intermediate in peptidoglycan synthesis. It is a substrate for peptidoglycan glycosyltransferase (EC 2.4.1.129) which catalyzes the chemical reaction [GlcNAc-(1->4)-Mur2Ac(oyl-L-Ala-gamma-D-Glu-L-Lys-D-Ala-D-Ala)]n- diphosphoundecaprenol + GlcNAc-(1->4)-Mur2Ac(oyl-L-Ala-gamma-D-Glu-L-Lys-D-Ala-D-Ala)- diphosphoundecaprenol <=> [GlcNAc-(1->4)-Mur2Ac(oyl-L-Ala-gamma-D-Glu-L-Lys-D-Ala-D-Ala)]n+1- diphosphoundecaprenol + undecaprenyl diphosphate.
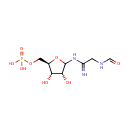
2-(Formamido)-N(1)-(5-phospho-D-ribosyl)acetamidine (PAMDB003416)
IUPAC:
{[(2R,3S,4R)-3,4-dihydroxy-5-(2-formamidoethanimidamido)oxolan-2-yl]methoxy}phosphonic acid
CAS: 6157-85-3
Description: 2-(Formamido)-N1-(5-phospho-D-ribosyl)acetamidine is an intermediate in purine metabolism. The enzyme phosphoribosylformylglycinamidine synthase [EC:6.3.5.3] catalyzes the production of this metabolite from N2-formyl-N1-(5-phospho-D-ribosyl)glycinamide.
Mur2Ac(oyl-L-Ala-gamma-D-Glu-L-Lys-D-Ala-D-Ala)-diphosphoundecaprenol (PAMDB003420)
IUPAC:
(2R)-4-{[(1S)-5-amino-1-{[(1S)-1-{[(1R)-1-carboxyethyl]-C-hydroxycarbonimidoyl}ethyl]-C-hydroxycarbonimidoyl}pentyl]-C-hydroxycarbonimidoyl}-2-{[(2S)-1-hydroxy-2-{[(2R)-1-hydroxy-2-{[(2R,3S,4R,5R,6R)-3-hydroxy-6-{[hydroxy({[hydroxy({[(2Z,6Z,10Z,14Z,18Z,22Z,26Z,30Z,34E,38E)-3,7,11,15,19,23,27,31,35,39,43-undecamethyltetratetraconta-2,6,10,14,18,22,26,30,34,38,42-undecaen-1-yl]oxy})phosphoryl]oxy})phosphoryl]oxy}-5-[(1-hydroxyethylidene)amino]-2-(hydroxymethyl)oxan-4-yl]oxy}propylidene]amino}propylidene]amino}butanoic acid
CAS: Not Available
Description: Mur2Ac(oyl-L-Ala-gamma-D-Glu-L-Lys-D-Ala-D-Ala)-diphosphoundecaprenol is an intermediate in peptidoglycan synthesis. It is a substrate for the enzyme undecaprenyldiphospho-muramoylpentapeptide beta-N-acetylglucosaminyltransferase which catalyzes the reaction: UDP-N-acetylglucosamine + Mur2Ac(oyl-L-Ala-gamma-D-Glu-L-Lys-D-Ala-D-Ala)-diphosphoundecaprenol = UDP + N-acetylglucosamine-(1-4)-Mur2Ac(oyl-L-Ala-gamma-D-Glu-L-Lys-D-Ala-D-Ala)-diphosphoundecaprenol.