Search Results for compounds
Searching compounds for
returned 4373 results.
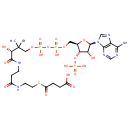
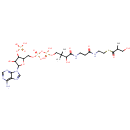
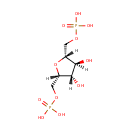
Succinyl-CoA (PAMDB000228)
IUPAC:
4-[(2-{3-[(2R)-3-[({[({[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)oxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy](hydroxy)phosphoryl}oxy)methyl]-2-hydroxy-3-methylbutanamido]propanamido}ethyl)sulfanyl]-4-oxobutanoic acid
CAS: 604-98-8
Description: Succinyl-CoA is a combination of succinic acid and coenzyme A. It is an important intermediate in the citric acid cycle, where it is synthesized from alpha-Ketoglutarate by alpha-ketoglutarate dehydrogenase through decarboxylation. During the process, coenzyme A is added. It is also synthesized from propionyl CoA, the odd-numbered fatty acid, which cannot undergo beta-oxidation. Propionyl-CoA is carboxylated to D-methylmalonyl-CoA, isomerized to L-methylmalonyl-CoA, and rearranged to yield succinyl-CoA via a vitamin B12-dependent enzyme. Succinyl-CoA is an intermediate of the citric acid cycle.
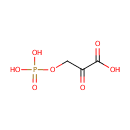
Phosphohydroxypyruvic acid (PAMDB000229)
IUPAC:
2-oxo-3-(phosphonooxy)propanoic acid
CAS: 3913-50-6
Description: Phosphohydroxypyruvic acid is a prduct of both enzyme phosphoglycerate dehydrogenase [EC 1.1.1.95] and phosphoserine transaminase [EC 2.6.1.52] in glycine, serine and threonine metabolism pathway (KEGG).
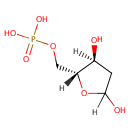
Deoxyribose 5-phosphate (PAMDB000230)
IUPAC:
{[(2R,3S)-3,5-dihydroxyoxolan-2-yl]methoxy}phosphonic acid
CAS: 102916-66-5
Description: Deoxyribose 5-phosphate is a a metabolite in the pentose phosphate pathway. It can be generated from D-glyceraldehdye-3 phosphate via the enzyme 2-Deoxyribose 5-phosphate aldolase (DERA). Alternately Deoxyribose 5-phosphate can be converted to D-glyceraldehyde-3 phosphate that can then feed into the pentose phosphate pathway. Deoxyribose 5-phosphate can also be generated from 2-Deoxy-D-ribose via the enzyme Ribokinase (EC 2.7.1.15). It has been shown in a number of organisms that deoxynucleosides or deoxyriboses cause the induction of aldolases (such as DERA) involved in their catabolism, leading to the utilisation of the pentose moiety as carbon and energy source.
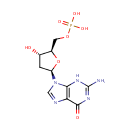
2'-Deoxyguanosine 5'-monophosphate (PAMDB000231)
IUPAC:
{[(2R,3S,5R)-5-(2-amino-6-oxo-6,9-dihydro-3H-purin-9-yl)-3-hydroxyoxolan-2-yl]methoxy}phosphonic acid
CAS: 902-04-5
Description: 2'-Deoxyguanosine 5'-monophosphate is a derivative of the common nucleic acid GTP, or guanosine triphosphate, in which the -OH (hydroxyl) group on the 2' carbon on the nucleotide's pentose has been removed (hence the deoxy- part of the name). Additionally, the diphosphate of the name indicates that two of the phosphoryl groups of GTP have been removed, most likely by hydrolysis (Wikipedia).
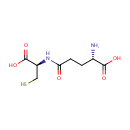
gamma-Glutamylcysteine (PAMDB000232)
IUPAC:
(2S)-2-amino-4-{[(1R)-1-carboxy-2-sulfanylethyl]carbamoyl}butanoic acid
CAS: 636-58-8
Description: G-Glutamylcysteine is a product of enzyme glutamate-cysteine ligase [EC 6.3.2.2] and a substrate of enzyme glutathione synthase [EC 6.3.2.3] in glutamate metabolism pathway (KEGG).
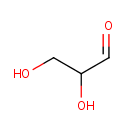
D-Glyceraldehyde (PAMDB000233)
IUPAC:
2,3-dihydroxypropanal
CAS: 56-82-6
Description: Glyceraldehyde is a triose monosaccharide with chemical formula C3H6O3. It is the simplest of all common aldoses. It is a sweet colorless crystalline solid that is an intermediate compound in carbohydrate metabolism. The word comes from combining glycerine and aldehyde, as glyceraldehyde is merely glycerine with one hydroxide changed to an aldehyde.
(S)-3-Hydroxyisobutyryl-CoA (PAMDB000234)
IUPAC:
{[5-(6-amino-9H-purin-9-yl)-4-hydroxy-2-({[hydroxy({[hydroxy(3-hydroxy-3-{[2-({2-[(3-hydroxy-2-methylpropanoyl)sulfanyl]ethyl}carbamoyl)ethyl]carbamoyl}-2,2-dimethylpropoxy)phosphoryl]oxy})phosphoryl]oxy}methyl)oxolan-3-yl]oxy}phosphonic acid
CAS: 319440-43-2
Description: (S)-3-Hydroxyisobutyryl-CoA is s metabolite of 3-hydroxyisobutyryl-CoA hydrolase (EC 3.1.2.4 ) during beta-alanine metabolism (KEGG 00410), propanoate metabolism (KEGG 00640), and valine, leucine and isoleucine degradation (KEGG 00280). Deficiencies of this enzyme in valine degradation can result in hypotonia, poor feeding, motor delay, and subsequent neurological regression in infancy, episodes of ketoacidosis and Leigh-like changes in the basal ganglia on a magnetic resonance imaging scan (PMID 17160907).
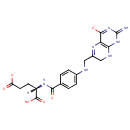
Dihydrofolic acid (PAMDB000235)
IUPAC:
(4S)-4-carboxy-4-[(4-{[(2-imino-4-oxido-1,2,7,8-tetrahydropteridin-6-yl)methyl]amino}phenyl)formamido]butanoate
CAS: 4033-27-6
Description: Dihydrofolic acid is a folic acid derivative acted upon by dihydrofolate reductase to produce tetrahydrofolic acid. It interacts with bacteria during cell division. It can be targeted with drug analogs to prevent nucleic acid synthesis. Dihydrofolic acid is also known by the name Dihydrofolate - more commonly Vitamin B9.
Fructose 1,6-bisphosphate (PAMDB000236)
IUPAC:
{[(2S,3S,4S,5R)-3,4-dihydroxy-5-[(phosphonooxy)methyl]oxolan-2-yl]methoxy}phosphonic acid
CAS: 488-69-7
Description: The hydrolysis of fructose 1,6-bisphosphate is catalized by Fructose-1,6-bisphosphatase (fru-1,6-P2ase, EC 3.1.3.11) to fructose 6-phosphate and inorganic phosphate and provides a mechanism to permit the reversal of the glycolysis reaction (catalyzed by 6-phosphofructo-1-kinase).(OMIM)
Ubiquinol-8 (PAMDB000237)
IUPAC:
2,3-dimethoxy-5-methyl-6-(3,7,11,15,19,23,27,31-octamethyldotriaconta-2,6,10,14,18,22,26,30-octaen-1-yl)cyclohexa-2,5-diene-1,4-diol
CAS: Not Available
Description: Ubiquinol-8 is a member of the chemical class known as Polyprenylbenzoquinols. They are reduced forms of polyprenylbenzoquinines (ubiquinones). These are compounds containing a polyisoprene chain attached to a quinol at the second ring position. Ubiquiol-1 has 8 isoprene units. Normally in Pseudomonas aeruginosa the active form of Ubiquinol has 8 isoprene units (Ubiquinol-8) and in humans it normally has 10. Coenzyme Q(n) exists in three redox states, fully oxidized (ubiquinone), partially reduced (semiquinones or ubisemiquinones), and fully reduced (ubiquinols). The redox functions of ubiquinol in cellular energy production and antioxidant protection are based on the ability to exchange two electrons in a redox cycle between ubiquinol (reduced) and the ubiquinone (oxidized) form. Ubiquionols are important in cellular respiration. They are fat-soluble and therefore mobile in cellular membranes; they play a unique role in the electron transport chain (ETC). In the inner bacterial membrane, electrons from NADH and succinate pass through the ETC to the oxygen, which is then reduced to water. The transfer of electrons through ETC results in the pumping of H+ across the membrane creating a proton gradient across the membrane, which is used by ATP synthase (located on the membrane) to generate ATP.