| Identification |
| Name: |
3-dehydroquinate synthase |
| Synonyms: |
Not Available |
| Gene Name: |
aroB |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in 3-dehydroquinate synthase activity |
| Specific Function: |
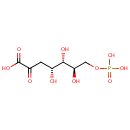
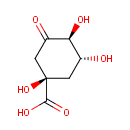
3-deoxy-D-arabino-hept-2-ulosonate 7-phosphate = 3-dehydroquinate + phosphate |
| Cellular Location: |
Cytoplasm |
| KEGG Pathways: |
- Metabolic pathways pae01100
- Phenylalanine, tyrosine and tryptophan biosynthesis pae00400
|
| KEGG Reactions: |
|
| SMPDB Reactions: |
|
| PseudoCyc/BioCyc Reactions: |
|
| Complex Reactions: |
|
| Transports: |
Not Available |
| Metabolites: |
| PAMDB ID | Name | View |
|---|
| PAMDB000888 | 2-Dehydro-3-deoxy-D-arabino-heptonate 7-phosphate | MetaboCard | | PAMDB000582 | 3-Dehydroquinate | MetaboCard | | PAMDB006299 | 3-deoxy-D-arabino-heptulosonate-7-phosphate | MetaboCard | | PAMDB001776 | Inorganic phosphate | MetaboCard | | PAMDB000383 | Phosphate | MetaboCard |
|
| GO Classification: |
| Component |
|---|
| cell part | | cytoplasm | | intracellular part | | Function |
|---|
| 3-dehydroquinate synthase activity | | carbon-oxygen lyase activity | | carbon-oxygen lyase activity, acting on phosphates | | catalytic activity | | lyase activity | | Process |
|---|
| aromatic amino acid family biosynthetic process | | biosynthetic process | | cellular amino acid and derivative metabolic process | | cellular amino acid biosynthetic process | | cellular amino acid metabolic process | | cellular metabolic process | | metabolic process |
|
| Gene Properties |
| Locus tag: |
PA5038 |
| Strand: |
- |
| Entrez Gene ID: |
881253 |
| Accession: |
NP_253725.1 |
| GI: |
15600231 |
| Sequence start: |
5674028 |
| Sequence End: |
5675134 |
| Sequence Length: |
1106 |
| Gene Sequence: |
>PA5038
ATGCGCACACTTCACGTCGATCTAGGCGAGCGTAGCTATCCCATTTACATCGGTGAAAACCTGCTGGGCGACGCCCGCTGGTTCGCGCCGCATATCGTCGGTCGCCGGGTCGCGGTGATCAGCAACGAAACGGTCGCTCCTCTCTATCTGGAAACCCTGCTCAAGGCACTGCAAGGCCACGAGGTCACCCCTGTGGTGCTGCCCGACGGCGAGGCCTACAAGCAGTGGGAAACCCTGCAACTGATCTTCGATGTCCTGCTGAAGGAGCGGCATGACCGCAAGACTACCCTGATCGCCCTGGGCGGCGGCGTCATCGGTGACATGGCCGGTTTCGCCGCGGCCTGCTACCAGCGTGGGGTCAACTTCATCCAGGTGCCGACCACCCTGTTGTCCCAGGTCGACTCCTCGGTGGGCGGCAAGACCGGGATCAACCATCCGTTGGGCAAGAACATGATCGGTGCGTTCTACCAGCCCCAGGCGGTGGTCATCGACACCGCGTCGCTGAAGACGCTGCCGTCCCGCGAGCTGTCCGCCGGGCTGGCGGAGGTGATCAAGTACGGTTTCATTTGCGACGAACCGTTCATCACCTGGCTGGAAGCGCACATGGATGCCTTGCTGGCACTGGAGCCGACCGTTGTCACCGAGGCCATCGAGCGCTCTTGCGCGGCGAAGGCCCGCGTGGTGGGCGCCGACGAGCGCGAGTCGGGCGTGCGTGCCACGCTGAACCTCGGCCATACCTTCGGTCACGCGATCGAGACCCAGCAAGGTTACGGCGTCTGGTTGCACGGCGAAGCGGTAGGGGCGGGAACCGTGATGGCACTCGAGATGTCGCATCGGTTGGGTTGGCTGAGCGCCGCCGAGCGCGATCGCGGTATCCGTCTGCTGCGCCGGGCTGGCCTGCCCGTGGTACCGCCGGCCGAGATGACGGCGGAAGATTTTATGGAACACATGGCCGTCGACAAGAAAGTGCTCGATGGCCGCCTGCGTCTGGTTTTGCTGCAAGGACTGGGGAATGCCGTGGTCACCGGCGATTTTCCCCGTGAGATCTTGGACGCGACCCTGCGCACCGACTATCGGGCCTTGGCCGACCAGCTTGGAGATGAATGA |
| Protein Properties |
| Protein Residues: |
368 |
| Protein Molecular Weight: |
40.1 kDa |
| Protein Theoretical pI: |
5.59 |
| Hydropathicity (GRAVY score): |
0.052 |
| Charge at pH 7 (predicted): |
-8.43 |
| Protein Sequence: |
>PA5038
MRTLHVDLGERSYPIYIGENLLGDARWFAPHIVGRRVAVISNETVAPLYLETLLKALQGHEVTPVVLPDGEAYKQWETLQLIFDVLLKERHDRKTTLIALGGGVIGDMAGFAAACYQRGVNFIQVPTTLLSQVDSSVGGKTGINHPLGKNMIGAFYQPQAVVIDTASLKTLPSRELSAGLAEVIKYGFICDEPFITWLEAHMDALLALEPTVVTEAIERSCAAKARVVGADERESGVRATLNLGHTFGHAIETQQGYGVWLHGEAVGAGTVMALEMSHRLGWLSAAERDRGIRLLRRAGLPVVPPAEMTAEDFMEHMAVDKKVLDGRLRLVLLQGLGNAVVTGDFPREILDATLRTDYRALADQLGDE |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA5038 and its homologs
|