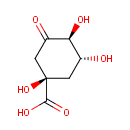
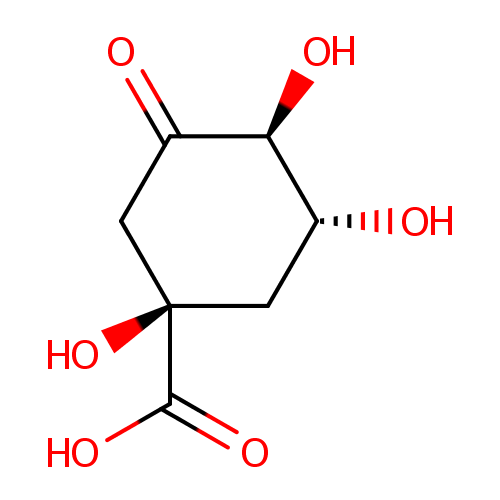
3-Dehydroquinate (PAMDB000582)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000582 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | 3-Dehydroquinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | 3-dehydroquinate is a member of the chemical class known as Cyclitols and Derivatives. These are compounds containing a cycloalkane moiety with one hydroxyl group on each of three or more ring atoms. 3-dehydroquinate is invovled in phenylalanine, tyrosine and tryptophan biosynthesis, and the biosynthesis of secondary metabolites. Dehydroquinate is involved in the shikimate pathway. The aroB-encoded enzyme dehydroquinate synthase is the second enzyme of this pathway, and it catalyzes the cyclization of 3-deoxy-D-arabino-heptulosonate-7-phosphate in 3-dehydroquinate. The aroB-encoded enzyme dehydroquinate synthase is the second enzyme of this pathway, and it catalyzes the cyclization of 3-deoxy-D-arabino-heptulosonate-7-phosphate in 3-dehydroquinate. (PMID 17586643) The aroB gene encoding dehydroquinate synthase (DHQS), which is a potential antibiotic target, was identified from the plant-pathogenic bacterium Xanthomonas oryzae pv. (PMID 19052366) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C7H10O6 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 190.1507 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 190.047738052 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | WVMWZWGZRAXUBK-SYTVJDICSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C7H10O6/c8-3-1-7(13,6(11)12)2-4(9)5(3)10/h3,5,8,10,13H,1-2H2,(H,11,12)/t3-,5+,7-/m1/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | (1R,3R,4S)-1,3,4-trihydroxy-5-oxocyclohexane-1-carboxylic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | 3-dehydroquinic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | O[C@@H]1C[C@@](O)(CC(=O)[C@H]1O)C(O)=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as cyclitols and derivatives. These are compounds containing a cycloalkane moiety with one hydroxyl group on each of three or more ring atoms. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organooxygen compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Alcohols and polyols | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Cyclic alcohols and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Cyclitols and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic homomonocyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | NAD + Quinate <> 3-Dehydroquinate +2 Hydrogen ion + NADH Quinate + NADP <> 3-Dehydroquinate + NADPH + Hydrogen ion 3-Dehydroquinate <> Water + 3-Dehydro-shikimate 2-Dehydro-3-deoxy-D-arabino-heptonate 7-phosphate > Phosphate + 3-Dehydroquinate NAD(P)<sup>+</sup> + Quinate NAD(P)H + 3-Dehydroquinate + Hydrogen ion 2-Dehydro-3-deoxy-D-arabino-heptonate 7-phosphate > 3-Dehydroquinate + Inorganic phosphate Quinate + NAD(P)(+) > 3-Dehydroquinate + NAD(P)H Quinate + NAD + NADP + Shikimic acid <> 3-Dehydroquinate + NADH + NADPH + Hydrogen ion + 3-Dehydro-shikimate 3-deoxy-D-arabino-heptulosonate-7-phosphate > Phosphate + 3-Dehydroquinate 3-Dehydroquinate > Water + 3-dehydroshikimate + 3-Dehydro-shikimate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||