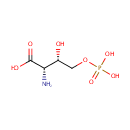| Identification |
| Name: |
Phosphoserine aminotransferase |
| Synonyms: |
- Phosphohydroxythreonine aminotransferase
- PSAT
|
| Gene Name: |
serC |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in metabolic process |
| Specific Function: |
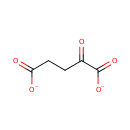
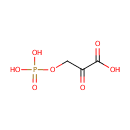
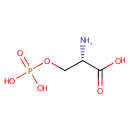
Catalyzes the reversible conversion of 3- phosphohydroxypyruvate to phosphoserine and of 3-hydroxy-2-oxo-4- phosphonooxybutanoate to phosphohydroxythreonine. Is involved in both pyridoxine and serine biosynthesis |
| Cellular Location: |
Cytoplasm |
| KEGG Pathways: |
|
| KEGG Reactions: |
|
| SMPDB Reactions: |
| | |
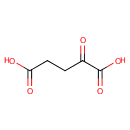
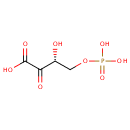 | + | L-Glutamic acid | + | 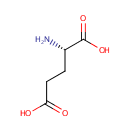 | ↔ | A-Ketoglutaric acid oxime | + | 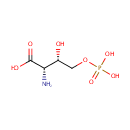 |
| | | | |
L-Glutamic acid | + |  | ↔ | 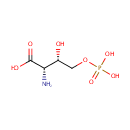 |
| | | | |
L-Glutamic acid | + |  | ↔ | 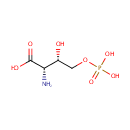 | + | A-Ketoglutaric acid oxime |
| | | |
|
| PseudoCyc/BioCyc Reactions: |
|
| Complex Reactions: |
Not Available |
| Transports: |
Not Available |
| Metabolites: |
|
| GO Classification: |
| Function |
|---|
| binding | | catalytic activity | | cofactor binding | | pyridoxal phosphate binding | | Process |
|---|
| cellular amino acid and derivative metabolic process | | cellular amino acid metabolic process | | cellular metabolic process | | L-serine biosynthetic process | | L-serine metabolic process | | metabolic process | | serine family amino acid metabolic process |
|
| Gene Properties |
| Locus tag: |
PA3167 |
| Strand: |
- |
| Entrez Gene ID: |
882700 |
| Accession: |
NP_251857.1 |
| GI: |
15598363 |
| Sequence start: |
3555254 |
| Sequence End: |
3556339 |
| Sequence Length: |
1085 |
| Gene Sequence: |
>PA3167
GTGAGCAAGCGAGCCTTCAATTTCTGCGCCGGTCCCGCGGCGCTTCCCGACGCAGTTCTGCAACGTGCCCAGGCCGAGCTTCTCGACTGGCGGGGCAAGGGCCTTTCGGTCATGGAAATGAGCCACCGTAGTGATGACTACGTGGCCATCGCCAGCAAGGCCGAGCAGGACCTGCGCGACCTGCTCGACATTCCCTCGGACTACAAGGTGCTGTTCCTGCAGGGCGGCGCCAGCCAGCAGTTCGCCGAGATTCCGCTGAACCTGCTGCCCGAAGATGGCGTCGCCGACTATATCGATACCGGTATCTGGTCGAAGAAGGCCATCGAGGAGGCTCGTCGCTATGGCACCGTGAATGTCGCGGCCAGCGCCAAGGAGTACGACTACTTCGCCATTCCGGGTCAGAACGAGTGGACGCTGACCAAGGACGCGGCCTATGTGCACTACGCCTCCAACGAGACCATCGGCGGCCTCGAGTTCGACTGGATTCCCGAGACCGGCGACGTGCCGCTGGTCACCGACATGTCCTCCGACATCCTCTCGCGCCCGCTCGACGTGTCCCGCTTCGGCCTGATCTATGCCGGGGCGCAGAAGAACATCGGCCCGTCCGGCCTGGTGGTGGTGATCGTTCGCGAAGACCTGCTCGGCCGCGCCCGCAGCGTCTGTCCGACCATGCTCAACTACAAGACCGCCGCGGACAACGGTTCCATGTACAACACCCCGGCGACTTACTCCTGGTACCTGTCCGGCCTGGTCTTCGAATGGCTGAAGGAGCAGGGCGGGGTGACCGCGATGGAGCAGCGCAACCGCGCCAAGAAGGACCTGCTGTACAAGACCATCGATGCCAGCGACTTCTACACCAACCCGATCCAGCCGAGCGCCCGCTCCTGGATGAACGTGCCGTTCCGCCTGGCCGACGAGCGTCTCGACAAGCCGTTCCTGGAAGGCGCCGAGGCGCGCGGGCTGCTCAACCTGAAAGGCCACCGCTCGGTCGGCGGCATGCGCGCTTCCATCTACAACGCCCTTGGCCTGGACGCGGTCGAGGCGCTGGTCGCATACATGGCGGAGTTCGAGAAGGAGCACGGCTGA |
| Protein Properties |
| Protein Residues: |
361 |
| Protein Molecular Weight: |
39.9 kDa |
| Protein Theoretical pI: |
4.71 |
| Hydropathicity (GRAVY score): |
-0.232 |
| Charge at pH 7 (predicted): |
-11.09 |
| Protein Sequence: |
>PA3167
MSKRAFNFCAGPAALPDAVLQRAQAELLDWRGKGLSVMEMSHRSDDYVAIASKAEQDLRDLLDIPSDYKVLFLQGGASQQFAEIPLNLLPEDGVADYIDTGIWSKKAIEEARRYGTVNVAASAKEYDYFAIPGQNEWTLTKDAAYVHYASNETIGGLEFDWIPETGDVPLVTDMSSDILSRPLDVSRFGLIYAGAQKNIGPSGLVVVIVREDLLGRARSVCPTMLNYKTAADNGSMYNTPATYSWYLSGLVFEWLKEQGGVTAMEQRNRAKKDLLYKTIDASDFYTNPIQPSARSWMNVPFRLADERLDKPFLEGAEARGLLNLKGHRSVGGMRASIYNALGLDAVEALVAYMAEFEKEHG |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA3167 and its homologs
|