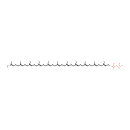| Identification |
| Name: |
Penicillin-binding protein 1C |
| Synonyms: |
- PBP-1c
- PBP1c
- Penicillin-insensitive transglycosylase
- Peptidoglycan TGase
- Transpeptidase-like module
|
| Gene Name: |
pbpC |
| Enzyme Class: |
Not Available |
| Biological Properties |
| General Function: |
Involved in catalytic activity |
| Specific Function: |
Cell wall formation. The enzyme has a penicillin- insensitive transglycosylase N-terminal domain (formation of linear glycan strands) and a transpeptidase C-terminal domain which may not be functional |
| Cellular Location: |
Cell inner membrane; Single-pass type II membrane protein |
| KEGG Pathways: |
|
| KEGG Reactions: |
Not Available |
| SMPDB Reactions: |
|
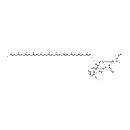
2  | → | 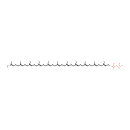 | + |  | + | a peptidoglycan dimer (meso-diaminopimelate containing) |
| | |
2 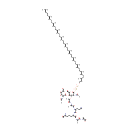 | → | a peptidoglycan dimer (meso-diaminopimelate containing) | + | 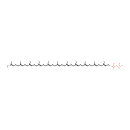 | + |  |
| |
|
| PseudoCyc/BioCyc Reactions: |
|
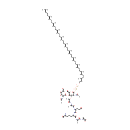
 | → |  | + | a peptidoglycan dimer (meso-diaminopimelate containing) | + | 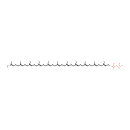 |
| |
|
| Complex Reactions: |
|
 | + | two linked disacharide pentapeptide murein units (uncrosslinked, middle of chain) | → |  | + | 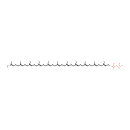 | + | three linked disacharide pentapeptide murein units (uncrosslinked, middle of chain) |
| | |
2  | → | 2  | + | 2  | + | two linked disacharide pentapeptide murein units (uncrosslinked, middle of chain) |
| |
|
| Transports: |
Not Available |
| Metabolites: |
| PAMDB ID | Name | View |
|---|
| PAMDB001633 | Hydrogen ion | MetaboCard | | PAMDB003413 | lipid II(A) | MetaboCard | | PAMDB001129 | N-Acetylmuramoyl-L-alanyl-D-glutamyl-L-lysyl-D-alanyl-D-alanine-diphosphoundecaprenyl-N-acetylglucosamine | MetaboCard | | PAMDB000394 | Undecaprenyl diphosphate | MetaboCard | | PAMDB001056 | Undecaprenyl-diphospho-N-acetylmuramoyl-(N-acetylglucosamine)-L-alanyl-D-glutaminyl-meso-2,6-diaminopimeloyl-D-alanyl-D-alanine | MetaboCard | | PAMDB001058 | Undecaprenyl-diphospho-N-acetylmuramoyl-(N-acetylglucosamine)-L-alanyl-D-glutamyl-meso-2,6-diaminopimeloyl-D-alanyl-D-alanine | MetaboCard |
|
| GO Classification: |
| Component |
|---|
| cell part | | cell wall | | external encapsulating structure | | peptidoglycan-based cell wall | | Function |
|---|
| binding | | catalytic activity | | drug binding | | penicillin binding | | Process |
|---|
| aminoglycan metabolic process | | carbohydrate metabolic process | | cell wall biogenesis | | cell wall organization or biogenesis | | cellular cell wall organization or biogenesis | | cellular component organization or biogenesis | | glycosaminoglycan metabolic process | | metabolic process | | peptidoglycan biosynthetic process | | peptidoglycan metabolic process | | peptidoglycan-based cell wall biogenesis | | polysaccharide metabolic process | | primary metabolic process |
|
| Gene Properties |
| Locus tag: |
PA2272 |
| Strand: |
+ |
| Entrez Gene ID: |
882173 |
| Accession: |
NP_250962.1 |
| GI: |
15597468 |
| Sequence start: |
2501720 |
| Sequence End: |
2503417 |
| Sequence Length: |
1697 |
| Gene Sequence: |
>PA2272
ATGAGCAGTCAACGCCGAAACTACCGCTTCATCCTTGTCGTCACCCTGTTCGTCCTCGCCTCCCTGGCCGTCTCCGGACGGTTGGTCTATCTCCAGGTCCACGACCACGAATTCCTCGCCGACCAGGGCGACCTCCGCTCGATCCGCGACCTGCCGATCCCGGTCACCCGCGGCATGATCACCGACCGCAACGGCGAGCCGCTGGCGGTATCCACCGAAGTCGCGTCGATCTGGTGCAACCCCAGGGAAATGGCCGCCCACCTCGACGAGGTGCCGCGCCTGGCCGGCGCCCTGCACCGCCCCGCGGCGGCGCTGCTGGCCCAGCTCCAGGCCAACCCGAACAAGCGCTTCCTCTACCTCGAGCGCGGCCTGTCGCCGATCGAGGCCAGCGAGGTGATGGCCCTGGGCATAACGGGGGTACACCAGATCAAGGAATACAAGCGTTTCTACCCCAGTTCCGAGCTGACCGCGCAGTTGATCGGCCTGGTCAACATCGACGGCCGCGGCCAGGAAGGCACCGAACTGGGCTTCAACGACTGGCTGAGCGGCAAGGACGGGGTACGCGAGGTGGCGATCAACCCGCGCGGCTCGCTGGTCAACAGCATCAAGGTGCTGAAGACGCCCAAGGCCAGCCAGGACGTGGCCCTGAGCATCGACCTGCGACTACAGTTCATCGCCTACAAGGCGCTGGAAAAGGCCGTGCTCAAGTTCGGCGCGCACTCCGGCTCGGCGGTCCTGGTGAACCCGAAGAGCGGGCAGATCCTGGCGATGGCCAACTTCCCCTCCTACAACCCGAACAACCGCGCCAGCTTCGCCCCGGCCTTCATGCGCAACCGCACCCTCACCGATACCTTCGAGCCGGGCTCGGTGATCAAGCCGTTCAGCATGTCGGCGGCGCTGGCCTCCGGCAAGTTCGACGAGAACAGTCAAGTCAGCGTGGCACCGGGCTGGATGACCATCGACGGGCACACCATCCACGACGTCGCCCGGCGCGACGTACTGACCATGACCGGGGTGCTGATCAACTCCTCGAACATCGGCATGAGCAAGGTCGCCCTGCAGATCGGACCCAAGCCGATCCTCGAACAGCTCGGCCGGGTCGGTTTCGGCGCGCCGCTGTCGCTGGGCTTCCCCGGCGAGAACCCGGGCTACCTGCCGTTCCACGAGAAATGGTCGAACATCGCCACCGCCAGCATGTCGTTCGGCTACAGCCTGGCGGTGAACACCGCCGAGCTGGCCCAGGCCTACTCGGTGTTCGCCAACGACGGCAAGCTGGTGCCGCTCAGCCTGCTCCGCGACAACCCGCAGAACCAGGTGCGACAGGCGATGGACCCGCAGATCGCACGGCGCATCCGGGCGATGCTGCAAACCGTGGTGGAAGACCCGAAGGGCGTGGTCCGCGCCCGCGTGCCGGGCTACCACGTGGCGGGCAAGAGCGGCACCGCGCGCAAGGCCTCGGGCCGGGGCTACGCGGACAAGTCCTACCGTTCGCTGTTCGTCGGCATGGCGCCGGCGTCCGACCCGCAACTGGTGCTGGCGGTGATGATCGATTCGCCGACCAGGATCGGCTACTTCGGCGGCCTGGTCTCGGCGCCCACCTTCAGCGACATCATGGCCGGATCGCTACGCGCCCTGGCGATCCCGCCGGACAACCTGCAGGACAGCCCGGCCGTCGCCGACCGCCAGCACCACGGCTGA |
| Protein Properties |
| Protein Residues: |
565 |
| Protein Molecular Weight: |
61.1 kDa |
| Protein Theoretical pI: |
9.92 |
| Hydropathicity (GRAVY score): |
-0.058 |
| Charge at pH 7 (predicted): |
12.86 |
| Protein Sequence: |
>PA2272
MSSQRRNYRFILVVTLFVLASLAVSGRLVYLQVHDHEFLADQGDLRSIRDLPIPVTRGMITDRNGEPLAVSTEVASIWCNPREMAAHLDEVPRLAGALHRPAAALLAQLQANPNKRFLYLERGLSPIEASEVMALGITGVHQIKEYKRFYPSSELTAQLIGLVNIDGRGQEGTELGFNDWLSGKDGVREVAINPRGSLVNSIKVLKTPKASQDVALSIDLRLQFIAYKALEKAVLKFGAHSGSAVLVNPKSGQILAMANFPSYNPNNRASFAPAFMRNRTLTDTFEPGSVIKPFSMSAALASGKFDENSQVSVAPGWMTIDGHTIHDVARRDVLTMTGVLINSSNIGMSKVALQIGPKPILEQLGRVGFGAPLSLGFPGENPGYLPFHEKWSNIATASMSFGYSLAVNTAELAQAYSVFANDGKLVPLSLLRDNPQNQVRQAMDPQIARRIRAMLQTVVEDPKGVVRARVPGYHVAGKSGTARKASGRGYADKSYRSLFVGMAPASDPQLVLAVMIDSPTRIGYFGGLVSAPTFSDIMAGSLRALAIPPDNLQDSPAVADRQHHG |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA2272 and its homologs
|