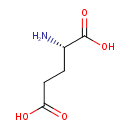
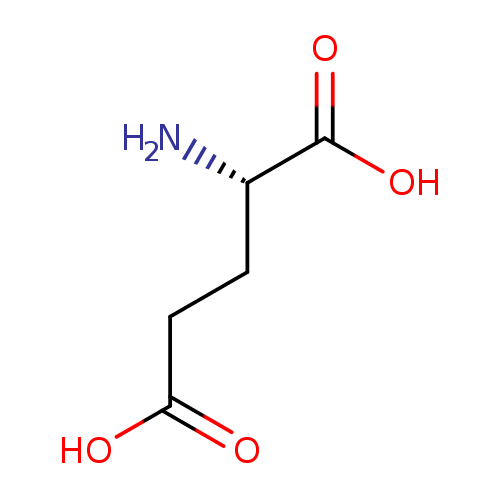
L-glutamate (PAMDB110666)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB110666 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | L-glutamate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | An α-amino-acid anion that is the conjugate base of L-glutamic acid, having anionic carboxy groups and a cationic amino group | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C5H8NO4 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 146.12 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 148.0609828146 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | WHUUTDBJXJRKMK-VKHMYHEASA-M | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C5H9NO4/c6-3(5(9)10)1-2-4(7)8/h3H,1-2,6H2,(H,7,8)(H,9,10)/p-1/t3-/m0/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 56-86-0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | (2S)-2-aminopentanedioic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | L-glutamic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | C(CCC(C(=O)[O-])[N+])([O-])=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as glutamic acid and derivatives. These are compounds containing glutamic acid or a derivative thereof resulting from reaction of glutamic acid at the amino group or the carboxy group, or from the replacement of any hydrogen of glycine by a heteroatom. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Carboxylic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Amino acids, peptides, and analogues | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Glutamic acid and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | L-glutamate + L-Tyrosine + L-2,4-diaminobutanoate + L-Serine + L-arginine + N5-formyl-N5-hydroxy-L-ornithine + L-lysine + L-Threonine → Hydrogen ion + ferribactin + Water L-Phenylalanine + alpha-Ketoglutarate → 2-oxo-3-phenylpropanoate + L-glutamate (S)-1-pyrroline-5-carboxylate + NAD+ + Water → Hydrogen ion + L-glutamate + NADH S-Substituted-Glutathione + Water → CPD-6262 + L-glutamate L-Glutamine + L-aspartate + ATP + Water → Hydrogen ion + L-glutamate + L-Asparagine + diphosphate + AMP Aromatic-Amino-Acids + alpha-Ketoglutarate → Aromatic-Oxoacids + L-glutamate alpha-Ketoglutarate + L-Alanine → L-glutamate + pyruvate L-glutamate + Water + NAD+ → alpha-Ketoglutarate + Ammonium + NADH + Hydrogen ion beta-Alanine + alpha-Ketoglutarate → 3-oxopropanoate + L-glutamate METHYLENE-THF-GLU-N + L-glutamate + ATP → METHYLENE-THF-GLU-N + ADP + phosphate 5-oxoproline + Water + ATP → Hydrogen ion + L-glutamate + phosphate + ADP alpha-N-Peptidyl-LGlutamate + L-glutamate + ATP → CPD0-2471 + ADP + phosphate + Hydrogen ion N-acetylglutaminylglutamine + L-Glutamine → N-acetylglutaminylglutamine amide + L-glutamate NAD-P-OR-NOP + Water + L-glutamate → NADH-P-OR-NOP + alpha-Ketoglutarate + Ammonium + Hydrogen ion L-Methionine + alpha-Ketoglutarate → 2-oxo-4-methylthiobutanoate + L-glutamate Uridine 5''-diphospho-{beta}-4-deoxy-4-amino-L-arabinose + alpha-Ketoglutarate → UDP-4-Keto-pyranose + L-glutamate alpha-Ketoglutarate + Aminated-Amine-Donors → L-glutamate + Deaminated-Amine-Donors Water + N-formyl-L-glutamate → L-glutamate + formate Hydrogen ion + L-glutamate → Carbon dioxide + gamma-Aminobutyric acid cob(II)yrinate c-monoamide + L-Glutamine + ATP + Water → cob(II)yrinate a,c-diamide + L-glutamate + ADP + phosphate + Hydrogen ion More...LysW-L-ornithine + alpha-Ketoglutarate → L-glutamate + LysW-L-glutamate-5-semialdehyde cobyrinate + L-Glutamine + ATP + Water → cob(II)yrinate c-monoamide + L-glutamate + ADP + phosphate + Hydrogen ion L-2,4-diaminobutanoate + alpha-Ketoglutarate → L-Aspartate-semialdehyde + L-glutamate cobyrinate + L-Glutamine + ATP + Water → cob(II)yrinate a,c-diamide + L-glutamate + ADP + phosphate + Hydrogen ion 4-hydroxy-2-nonenal-glutathione conjugate + Water → 4-hydroxy-2-nonenal-[Cys-Gly] conjugate + L-glutamate L-Glutamine + ATP + Water → L-glutamate + phosphate + ADP + Hydrogen ion L-glutamate + ATP + Hydrogen ion → AMP + diphosphate L-glutamate + LysW-C-Terminal-L-Glutamate + ATP → LysW-L-glutamate + ADP + phosphate + Hydrogen ion tRNAs-Asp-with-queuosine + ATP + L-glutamate → tRNAs-with-glutamylated-queuosine + AMP + diphosphate + Hydrogen ion UDP-2-acetamido-3-amino-2,3-dideoxy-α-D-glucuronate + alpha-Ketoglutarate → UDP-2-acetamido-2-deoxy-α-D-ribo-hex-3-uluronate + L-glutamate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Horner, L.; Gross, A. Tertiary phosphines. IV. Use of phosphine imines in causing the introduction of primary amino groups. Ann. (1955), 591 117-34. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Download (PDF) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||