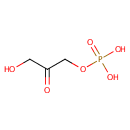
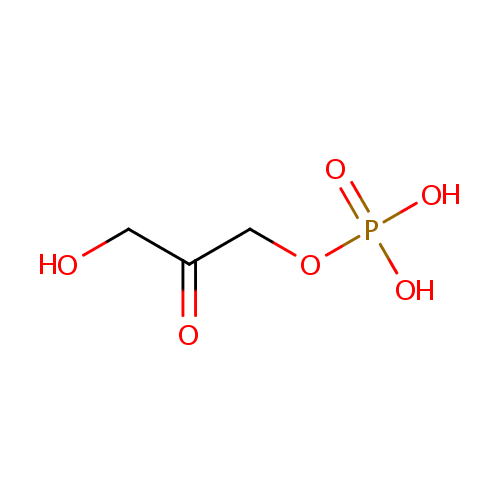
Dihydroxyacetone phosphate (PAMDB000395)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000395 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Dihydroxyacetone phosphate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | Dihydroxyacetone phosphate is integral to the glycolysis metabolic pathway, and is one of the two products of breakdown of fructose 1,6-bisphosphate, along with glyceraldehyde 3-phosphate. It is rapidly and reversibly isomerised to glyceraldehyde 3-phosphate. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C3H7O6P | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 170.0578 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 169.998024468 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | GNGACRATGGDKBX-UHFFFAOYSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C3H7O6P/c4-1-3(5)2-9-10(6,7)8/h4H,1-2H2,(H2,6,7,8) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 57-04-5 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | (3-hydroxy-2-oxopropoxy)phosphonic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | dihydroxyacetone-phosphate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | OCC(=O)COP(O)(O)=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as glycerone phosphates. These are organic compounds containing a glycerone moiety that carries a phosphate group at the O-1 or O-2 position. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organooxygen compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Carbonyl compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Ketones | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Glycerone phosphates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Glycerol 3-phosphate + Ubiquinone-8 > Dihydroxyacetone phosphate + Ubiquinol-8 D-Tagatose 1,6-bisphosphate <> Dihydroxyacetone phosphate + D-Glyceraldehyde 3-phosphate Dihydroxyacetone + Phosphoenolpyruvic acid > Dihydroxyacetone phosphate + Pyruvic acid Fructose 1,6-bisphosphate <> Dihydroxyacetone phosphate + D-Glyceraldehyde 3-phosphate Glycerol 3-phosphate + Menaquinone 8 > Dihydroxyacetone phosphate + Menaquinol 8 2-Demethylmenaquinone 8 + Glycerol 3-phosphate > 2-Demethylmenaquinol 8 + Dihydroxyacetone phosphate L-Fuculose 1-phosphate <> Dihydroxyacetone phosphate + Lactaldehyde + (S)-Lactaldehyde Dihydroxyacetone phosphate + Iminoaspartic acid <>2 Water + Phosphate + Quinolinic acid Dihydroxyacetone phosphate <> Pyruvaldehyde + Phosphate Sedoheptulose 1,7-bisphosphate <> Dihydroxyacetone phosphate + D-Erythrose 4-phosphate Glycerol 3-phosphate + NADP <> Dihydroxyacetone phosphate + Hydrogen ion + NADPH L-Rhamnulose 1-phosphate <> Dihydroxyacetone phosphate + Lactaldehyde + (S)-Lactaldehyde Dihydroxyacetone phosphate <> D-Glyceraldehyde 3-phosphate Glycerol 3-phosphate + NAD <> Dihydroxyacetone phosphate + NADH + Hydrogen ion Glycerol 3-phosphate + FAD <> Dihydroxyacetone phosphate + FADH2 Adenosine triphosphate + Dihydroxyacetone <> ADP + Dihydroxyacetone phosphate beta-D-Fructose 1,6-bisphosphate <> Dihydroxyacetone phosphate + D-Glyceraldehyde 3-phosphate L-Fuculose 1-phosphate <> Dihydroxyacetone phosphate + Lactaldehyde L-Rhamnulose 1-phosphate <> Dihydroxyacetone phosphate + Lactaldehyde More...Fructose 1-phosphate <> Dihydroxyacetone phosphate + D-Glyceraldehyde D-Ribulose-1-phosphate <> Glycolaldehyde + Dihydroxyacetone phosphate NAD(P)<sup>+</sup> + Glycerol 3-phosphate < NAD(P)H + Dihydroxyacetone phosphate + Hydrogen ion Glycerol 3-phosphate + a ubiquinone <> Dihydroxyacetone phosphate + a ubiquinol an electron-transfer-related quinone + Glycerol 3-phosphate <> an electron-transfer-related quinol + Dihydroxyacetone phosphate Glycerol 3-phosphate + a quinone > Dihydroxyacetone phosphate + a quinol Glycerol 3-phosphate + NAD(P)(+) > Dihydroxyacetone phosphate + NAD(P)H Dihydroxyacetone phosphate > Pyruvaldehyde + Inorganic phosphate Dihydroxyacetone phosphate + Iminoaspartic acid > Quinolinic acid +2 Water + Inorganic phosphate Glycerol 3-phosphate + Quinone <> Dihydroxyacetone phosphate + Hydroquinone Glycerol 3-phosphate + NAD + NADP <> Dihydroxyacetone phosphate + NADH + NADPH + Hydrogen ion 6-Deoxy-6-sulfo-D-fructose 1-phosphate <> Dihydroxyacetone phosphate + 2-Hydroxy-3-oxopropane-1-sulfonate Fructose 1,6-bisphosphate + Fructose 1,6-bisphosphate <> D-Glyceraldehyde 3-phosphate + Dihydroxyacetone phosphate + D-Glyceraldehyde 3-phosphate D-Glyceraldehyde 3-phosphate + D-Glyceraldehyde 3-phosphate <> Dihydroxyacetone phosphate D-tagatofuranose 1,6-bisphosphate > Dihydroxyacetone phosphate + D-Glyceraldehyde 3-phosphate + D-Glyceraldehyde 3-phosphate L-rhamnulose 1-phosphate + L-Rhamnulose 1-phosphate > Dihydroxyacetone phosphate + (S)-lactaldehyde + Lactaldehyde L-fuculose 1-phosphate + L-Fuculose 1-phosphate > Dihydroxyacetone phosphate + (S)-lactaldehyde + Lactaldehyde 3-hydroxy-2,4-pentanedione 5-phosphate + Coenzyme A + 3-hydroxy-2,4-pentanedione 5-phosphate > Acetyl-CoA + Dihydroxyacetone phosphate Dihydroxyacetone phosphate + Hydrogen ion + NADPH + NADPH > NADP + Glycerol 3-phosphate Glycerol 3-phosphate + Ubiquinone-1 > Dihydroxyacetone phosphate + Ubiquinol-1 Glycerol 3-phosphate + menaquinone-8 > Menaquinol 8 + Dihydroxyacetone phosphate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Ballou, Clinton E.; Fischer, Hermann O. L. The synthesis of dihydroxyacetone phosphate. Journal of the American Chemical Society (1956), 78 1659-61. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||