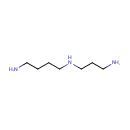
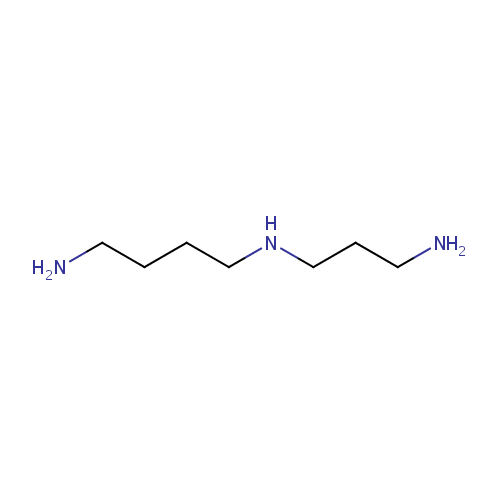
Spermidine (PAMDB000312)
| Record Information | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000312 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Spermidine | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | Spermidine is a polyamine formed from putrescine. It is found in almost all tissues in association with nucleic acids. It is found as a cation at all pH values, and is thought to help stabilize some membranes and nucleic acid structures. It is a precursor of spermine. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C7H19N3 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 145.2459 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 145.157897623 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | ATHGHQPFGPMSJY-UHFFFAOYSA-N | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C7H19N3/c8-4-1-2-6-10-7-3-5-9/h10H,1-9H2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 124-20-9 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | (4-aminobutyl)(3-aminopropyl)amine | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | spermidine | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | NCCCCNCCCN | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as dialkylamines. These are organic compounds containing a dialkylamine group, characterized by two alkyl groups bonded to the amino nitrogen. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organonitrogen compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Amines | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Secondary amines | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Dialkylamines | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Liquid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | 3 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | < 25 °C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Adenosine triphosphate + Water + Spermidine > ADP + Hydrogen ion + Phosphate + Spermidine Adenosine triphosphate + Water + Spermidine > ADP + Hydrogen ion + Phosphate + Spermidine S-Adenosylmethioninamine + Putrescine + Ethylenediamine <> 5'-Methylthioadenosine + Hydrogen ion + Spermidine Acetyl-CoA + Spermidine > N1-Acetylspermidine + Coenzyme A + Hydrogen ion Acetyl-CoA + Spermidine > Coenzyme A + Hydrogen ion + N8-Acetylspermidine Adenosine triphosphate + Glutathione + Spermidine <> ADP + Glutathionylspermidine + Hydrogen ion + Phosphate Glutathionylspermidine + Water <> Glutathione + Spermidine Adenosine triphosphate + Glutathione + Spermidine <> ADP + Phosphate + Glutathionylspermidine S-Adenosylmethioninamine + Putrescine <> 5'-Methylthioadenosine + Spermidine S-Adenosylmethioninamine + Spermidine <> 5'-Methylthioadenosine + Spermine -->-->Putrescine + S-Adenosylmethioninamine > Hydrogen ion + Spermidine + 5'-Methylthioadenosine Glutathione + Spermidine + Adenosine triphosphate > Glutathionylspermidine + ADP + Inorganic phosphate Putrescine + S-Adenosylmethioninamine > Spermidine + Hydrogen ion + 5'-S-methyl-5'-thioadenosine | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Bergeron, Raymond J., Jr. Preparation and formulation spermidine analogues for pharmaceutical use as tumor growth inhibitors. U.S. (2001), 31 pp. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Download (PDF) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in catalytic activity
- Specific function:
- Catalyzes the production of spermidine from putrescine and decarboxylated S-adenosylmethionine (dcSAM), which serves as an aminopropyl donor
- Gene Name:
- speE
- Locus Tag:
- PA1687
- Molecular weight:
- 32.2 kDa
Reactions
| S-adenosylmethioninamine + putrescine = 5'-S-methyl-5'-thioadenosine + spermidine. |
- General function:
- Involved in nucleotide binding
- Specific function:
- Part of the ABC transporter complex PotABCD involved in spermidine/putrescine import. Responsible for energy coupling to the transport system
- Gene Name:
- potA
- Locus Tag:
- PA3607
- Molecular weight:
- 40 kDa
Reactions
| ATP + H(2)O + polyamine(Out) = ADP + phosphate + polyamine(In). |
- General function:
- Involved in transporter activity
- Specific function:
- Required for the activity of the bacterial periplasmic transport system of putrescine and spermidine
- Gene Name:
- potB
- Locus Tag:
- PA3608
- Molecular weight:
- 32.7 kDa
- General function:
- Involved in transporter activity
- Specific function:
- Required for the activity of the bacterial periplasmic transport system of putrescine and spermidine
- Gene Name:
- potC
- Locus Tag:
- PA3609
- Molecular weight:
- 27.7 kDa
- General function:
- Involved in transporter activity
- Specific function:
- Required for the activity of the bacterial periplasmic transport system of putrescine and spermidine. Polyamine binding protein
- Gene Name:
- potD
- Locus Tag:
- PA3610
- Molecular weight:
- 39.3 kDa
Transporters
- General function:
- Involved in nucleotide binding
- Specific function:
- Part of the ABC transporter complex PotABCD involved in spermidine/putrescine import. Responsible for energy coupling to the transport system
- Gene Name:
- potA
- Locus Tag:
- PA3607
- Molecular weight:
- 40 kDa
Reactions
| ATP + H(2)O + polyamine(Out) = ADP + phosphate + polyamine(In). |
- General function:
- Involved in transporter activity
- Specific function:
- Required for the activity of the bacterial periplasmic transport system of putrescine and spermidine
- Gene Name:
- potB
- Locus Tag:
- PA3608
- Molecular weight:
- 32.7 kDa
- General function:
- Involved in transporter activity
- Specific function:
- Required for the activity of the bacterial periplasmic transport system of putrescine and spermidine
- Gene Name:
- potC
- Locus Tag:
- PA3609
- Molecular weight:
- 27.7 kDa
- General function:
- Involved in spermidine transmembrane transporter activity
- Specific function:
- Catalyzes the excretion of spermidine. Can also confer resistance to deoxycholate and SDS
- Gene Name:
- mdtI
- Locus Tag:
- PA1540
- Molecular weight:
- 11.8 kDa
- General function:
- Involved in spermidine transmembrane transporter activity
- Specific function:
- Catalyzes the excretion of spermidine. Can also confer resistance to deoxycholate and SDS
- Gene Name:
- mdtJ
- Locus Tag:
- PA1541
- Molecular weight:
- 12.7 kDa
- General function:
- Involved in transporter activity
- Specific function:
- Required for the activity of the bacterial periplasmic transport system of putrescine and spermidine. Polyamine binding protein
- Gene Name:
- potD
- Locus Tag:
- PA3610
- Molecular weight:
- 39.3 kDa