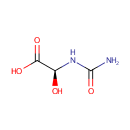
(S)-Ureidoglycolic acid (PAMDB000223)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000223 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | (S)-Ureidoglycolic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | (S)-Ureidoglycolic acid is a substrate of enzyme ureidoglycolate dehydrogenase [EC 1.1.1.154] in purine metabolism pathway (KEGG). Enteric bacteria such as Pseudomonas aeruginosa are able to utilize allantoin as a sole source of nitrogen under anaerobic conditions, but cannot utilize it as a sole source of carbon The first step in allantoin degradation is the opening of the aromatic ring, yielding allantoate, performed by allantoinase. In the next step allantoate is hydrolyzed to S-ureidoglycine by allantoate amidohydrolase. Ureidoglycine spontaneously converts to Ureidoglycolate. Ureidoglycolate dehydrogenase then oxidizes ureidoglycolate to oxalurate. It is believed oxalurate is converted into oxamate and carbamoyl-phosphate, which can be further metabolized to CO2, ammonia and ATP. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C3H6N2O4 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 134.0907 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 134.03275669 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | NWZYYCVIOKVTII-SFOWXEAESA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C3H6N2O4/c4-3(9)5-1(6)2(7)8/h1,6H,(H,7,8)(H3,4,5,9)/t1-/m0/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 7424-03-5 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | (2S)-2-(carbamoylamino)-2-hydroxyacetic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | ureidoglycolate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | NC(=O)N[C@@H](O)C(O)=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as n-carbamoyl-alpha amino acids. These are compounds containing an alpha amino acid which bears an carbamoyl group at its terminal nitrogen atom. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Carboxylic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Amino acids, peptides, and analogues | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | N-carbamoyl-alpha amino acids | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | 2 Hydrogen ion + Water + (S)-Ureidoglycolic acid > Carbon dioxide + Glyoxylic acid +2 Ammonium Allantoic acid + 2 Hydrogen ion + 2 Water > Carbon dioxide +2 Ammonium + (S)-Ureidoglycolic acid NAD + (S)-Ureidoglycolic acid <> Hydrogen ion + NADH + Oxalureate (S)-Ureidoglycolic acid + Water <> Glyoxylic acid +2 Ammonia + Carbon dioxide (S)-Ureidoglycolic acid + NADP <> Oxalureate + NADPH + Hydrogen ion Ureidoglycine + Water <> (S)-Ureidoglycolic acid + Ammonia NAD(P)<sup>+</sup> + (S)-Ureidoglycolic acid > NAD(P)H + Oxalureate + Hydrogen ion (S)-Ureidoglycolic acid > Urea + Glyoxylic acid <i>S</i>-ureidoglycine + Water > Hydrogen ion + Ammonia + (S)-Ureidoglycolic acid (S)-Ureidoglycolic acid + NAD(P)(+) > Oxalureate + NAD(P)H (S)-Ureidoglycolic acid + NAD + NADP <> Oxalureate + NADH + NADPH + Hydrogen ion S-ureidoglycine + Water > Ammonium + (S)-Ureidoglycolic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in ureidoglycolate hydrolase activity
- Specific function:
- Involved in the anaerobic utilization of allantoin. Reinforces the induction of genes involved in the degradation of allantoin and glyoxylate by producing glyoxylate
- Gene Name:
- allA
- Locus Tag:
- PA1514
- Molecular weight:
- 19 kDa
Reactions
| (S)-ureidoglycolate + H(2)O = glyoxylate + 2 NH(3) + CO(2). |