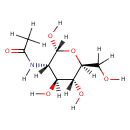
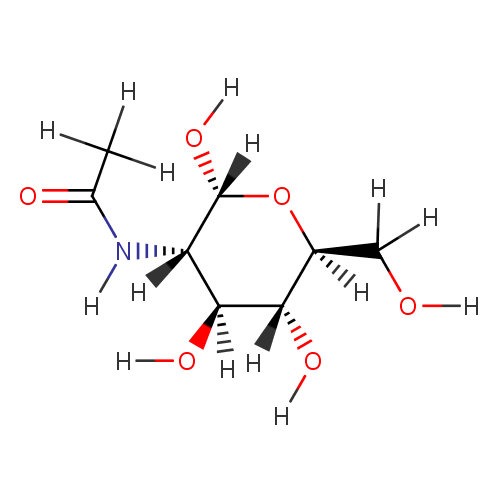
N-Acetyl-D-glucosamine (PAMDB000088)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000088 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | N-Acetyl-D-glucosamine | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | N-Acetyl-D-Glucosamine (N-acetlyglucosamine) is a monosaccharide derivative of glucose. Chemically it is an amide between glucosamine and acetic acid. A single N-acetlyglucosamine moiety linked to serine or threonine residues on nuclear and cytoplasmic proteins -O-GlcNAc, is an ubiquitous post-translational protein modification. O-GlcNAc modified proteins are involved in sensing the nutrient status of the surrounding cellular environment and adjusting the activity of cellular proteins accordingly. O-GlcNAc initiates a protective response to stress, modulates a cell's capacity to grow and divide, and regulates gene transcription. (PMID 16237703) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C8H15NO6 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 221.2078 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 221.089937217 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | OVRNDRQMDRJTHS-DMHSOCPYSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C8H15NO6/c1-3(11)9-5-7(13)6(12)4(2-10)15-8(5)14/h4-8,10,12-14H,2H2,1H3,(H,9,11)/t4-,5-,6-,7-,8+/m0/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 7512-17-6 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | N-[(2R,3S,4S,5R,6S)-2,4,5-trihydroxy-6-(hydroxymethyl)oxan-3-yl]acetamide | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | N-[(2R,3S,4S,5R,6S)-2,4,5-trihydroxy-6-(hydroxymethyl)oxan-3-yl]acetamide | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | [H]OC([H])([H])[C@]1([H])O[C@@]([H])(O[H])[C@@]([H])(N([H])C(=O)C([H])([H])[H])[C@]([H])(O[H])[C@@]1([H])O[H] | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as n-acyl-alpha-hexosamines. These are carbohydrate derivatives containing a hexose moiety in which the oxygen atom is replaced by an n-acyl group. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organooxygen compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Carbohydrates and carbohydrate conjugates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Aminosaccharides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | N-acyl-alpha-hexosamines | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic heteromonocyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | 0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | 210 °C | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Phosphoenolpyruvic acid + N-Acetyl-D-glucosamine > N-Acetyl-D-Glucosamine 6-Phosphate + Pyruvic acid N-Acetyl-D-glucosamine(anhydrous)N-Acetylmuramic acid + Water > N-Acetyl-D-glucosamine + 1,6-Anhydro-N-acetylmuramate N-Acetyl-D-glucosamine(anhydrous)N-Acetylmuramyl-tetrapeptide + Water > N-Acetyl-D-glucosamine + 1,6-Anhydrous-N-Acetylmuramyl-tetrapeptide N-Acetyl-D-glucosamine(anhydrous)N-Acetylmuramyl-tripeptide + Water > N-Acetyl-D-glucosamine + 1,6-Anhydrous-N-Acetylmuramyl-tripeptide N-Acetyl-D-glucosamine + Adenosine triphosphate <> N-Acetyl-D-Glucosamine 6-Phosphate + ADP + Hydrogen ion Diacetylchitobiose-6-phosphate + Water > N-Acetyl-D-glucosamine + N-Acetyl-D-Glucosamine 6-Phosphate Chitobiose + Water <>2 N-Acetyl-D-glucosamine Adenosine triphosphate + N-Acetyl-D-glucosamine <> ADP + N-Acetyl-D-Glucosamine 6-Phosphate Chitin + Water <> N-Acetyl-D-glucosamine + Chitin Protein N(pi)-phospho-L-histidine + N-Acetyl-D-glucosamine <> Protein histidine + N-Acetyl-D-Glucosamine 6-Phosphate a peptidoglycan + Water a peptidoglycan with cleaved N-acetyl-glucosamine + N-Acetyl-D-glucosamine <i>N</i>-acetyl-β-D-glucosamine(anhydrous)-<i>N</i>-acetylmuramate + Water N-Acetyl-D-glucosamine + 1,6-Anhydro-N-acetylmuramate Adenosine triphosphate + Water + N-Acetyl-D-glucosamine > N-Acetyl-D-glucosamine + ADP + Phosphate + Hydrogen ion Adenosine triphosphate + Water + N-Acetyl-D-glucosamine > N-Acetyl-D-glucosamine + ADP + Phosphate + Hydrogen ion | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Zhang, He; Qi, Shanlong; Yang, Shenggui. Production of N-acetyl-D-glucosamine from chitin. Faming Zhuanli Shenqing Gongkai Shuomingshu (2006), 6pp. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Download (PDF) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in protein-N(PI)-phosphohistidine-sugar phosphotransferase activity
- Specific function:
- The phosphoenolpyruvate-dependent sugar phosphotransferase system (sugar PTS), a major carbohydrate active -transport system, catalyzes the phosphorylation of incoming sugar substrates concomitantly with their translocation across the cell membrane. This system is involved in N-acetylglucosamine transport
- Gene Name:
- nagE
- Locus Tag:
- PA3761
- Molecular weight:
- 60.6 kDa
Reactions
| Protein EIIA N(pi)-phospho-L-histidine + protein EIIB = protein EIIA + protein EIIB N(pi)-phospho-L-histidine/cysteine. |
| Protein EIIB N(pi)-phospho-L-histidine/cysteine + sugar = protein EIIB + sugar phosphate. |
- General function:
- Involved in beta-N-acetylhexosaminidase activity
- Specific function:
- Cleaves GlcNAc linked beta-1,4 to MurNAc tripeptides
- Gene Name:
- nagZ
- Locus Tag:
- PA3005
- Molecular weight:
- 36.1 kDa
Reactions
| Hydrolysis of terminal non-reducing N-acetyl-D-hexosamine residues in N-acetyl-beta-D-hexosaminides. |
Transporters
- General function:
- Involved in protein-N(PI)-phosphohistidine-sugar phosphotransferase activity
- Specific function:
- The phosphoenolpyruvate-dependent sugar phosphotransferase system (sugar PTS), a major carbohydrate active -transport system, catalyzes the phosphorylation of incoming sugar substrates concomitantly with their translocation across the cell membrane. This system is involved in N-acetylglucosamine transport
- Gene Name:
- nagE
- Locus Tag:
- PA3761
- Molecular weight:
- 60.6 kDa
Reactions
| Protein EIIA N(pi)-phospho-L-histidine + protein EIIB = protein EIIA + protein EIIB N(pi)-phospho-L-histidine/cysteine. |
| Protein EIIB N(pi)-phospho-L-histidine/cysteine + sugar = protein EIIB + sugar phosphate. |