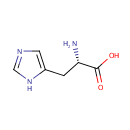
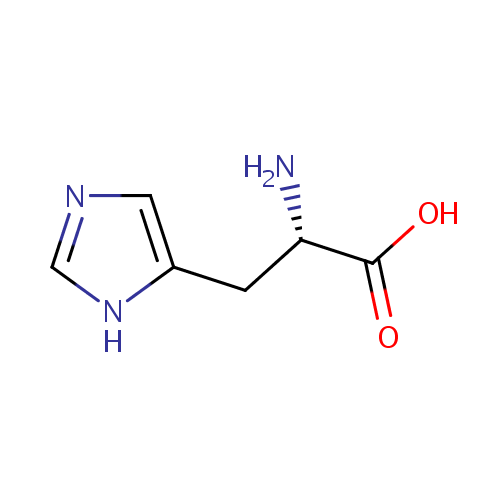
L-Histidine (PAMDB000074)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 12:54:54 PM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000074 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | L-Histidine | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | Histidine is an amino acid. It is a precursor for histamine and carnosine biosynthesis. The enzyme histidine ammonia-lyase converts histidine into ammonia and urocanic acid. (Wikipedia) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C6H9N3O2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 155.1546 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 155.069476547 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | HNDVDQJCIGZPNO-YFKPBYRVSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C6H9N3O2/c7-5(6(10)11)1-4-2-8-3-9-4/h2-3,5H,1,7H2,(H,8,9)(H,10,11)/t5-/m0/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 71-00-1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | (2S)-2-amino-3-(1H-imidazol-5-yl)propanoic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | L-histidine | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | N[C@@H](CC1=CN=CN1)C(O)=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as d-alpha-amino acids. These are alpha amino acids which have the D-configuration of the alpha-carbon atom. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Carboxylic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Amino acids, peptides, and analogues | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | D-alpha-amino acids | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic heteromonocyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | 0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | 287 °C | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Adenosine triphosphate + Water + L-Histidine > ADP + Hydrogen ion + L-Histidine + Phosphate Adenosine triphosphate + Water + L-Histidine > ADP + Hydrogen ion + L-Histidine + Phosphate Water + L-Histidinol + 2 NAD >3 Hydrogen ion + L-Histidine +2 NADH Adenosine triphosphate + L-Histidine + tRNA(His) + tRNA(His) <> Adenosine monophosphate + L-Histidyl-tRNA(His) + Pyrophosphate + L-Histidyl-tRNA(His) L-Histidinal + Water + NAD <> L-Histidine + NADH + Hydrogen ion Adenosine triphosphate + L-Histidine + tRNA(His) <> Adenosine monophosphate + Pyrophosphate + L-Histidyl-tRNA(His) -->-->histidinal + NAD + Water > Hydrogen ion + L-Histidine + NADH ala-his + Water > L-Alanine + L-Histidine L-Histidinol + Water + 2 NAD > L-Histidine +2 NADH L-Histidine + Adenosine triphosphate + Hydrogen ion + tRNA(His) + L-Histidine > Adenosine monophosphate + Pyrophosphate + L-histidyl-tRNA(His) Water + NAD + Histidinal >2 Hydrogen ion + NADH + L-Histidine + L-Histidine L-Histidine + Adenosine triphosphate + Water + L-Histidine > Adenosine diphosphate + Phosphate + Hydrogen ion + L-Histidine + ADP | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Aurelio Luigi; Brownlee Robert T C; Hughes Andrew B A novel synthesis of N-methyl asparagine, arginine, histidine, and tryptophan. Organic letters (2002), 4(21), 3767-9. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Download (PDF) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in oxidoreductase activity
- Specific function:
- Catalyzes the sequential NAD-dependent oxidations of L- histidinol to L-histidinaldehyde and then to L-histidine
- Gene Name:
- hisD
- Locus Tag:
- PA4448
- Molecular weight:
- 47.2 kDa
Reactions
| L-histidinol + H(2)O + 2 NAD(+) = L-histidine + 2 NADH. |
- General function:
- Involved in nucleotide binding
- Specific function:
- Part of the binding-protein-dependent transport system for histidine. Probably responsible for energy coupling to the transport system
- Gene Name:
- hisP
- Locus Tag:
- PA2926
- Molecular weight:
- 28.5 kDa
- General function:
- Involved in nucleotide binding
- Specific function:
- ATP + L-histidine + tRNA(His) = AMP + diphosphate + L-histidyl-tRNA(His)
- Gene Name:
- hisS
- Locus Tag:
- PA3802
- Molecular weight:
- 47.5 kDa
Reactions
| ATP + L-histidine + tRNA(His) = AMP + diphosphate + L-histidyl-tRNA(His). |
- General function:
- Involved in transporter activity
- Specific function:
- Part of the binding-protein-dependent transport system for histidine; probably responsible for the translocation of the substrate across the membrane
- Gene Name:
- hisM
- Locus Tag:
- PA2925
- Molecular weight:
- 26.7 kDa
- General function:
- Involved in transporter activity
- Specific function:
- Part of the binding-protein-dependent transport system for histidine; probably responsible for the translocation of the substrate across the membrane
- Gene Name:
- hisQ
- Locus Tag:
- PA2924
- Molecular weight:
- 24.5 kDa
- General function:
- Involved in transporter activity
- Specific function:
- Component of the high-affinity histidine permease, a binding-protein-dependent transport system. The other components are proteins hisQ, hisM, and hisP
- Gene Name:
- hisJ
- Locus Tag:
- PA2923
- Molecular weight:
- 28.2 kDa
Transporters
- General function:
- Involved in nucleotide binding
- Specific function:
- Part of the binding-protein-dependent transport system for histidine. Probably responsible for energy coupling to the transport system
- Gene Name:
- hisP
- Locus Tag:
- PA2926
- Molecular weight:
- 28.5 kDa
- General function:
- Involved in nucleotide binding
- Specific function:
- Probably part of a binding-protein-dependent transport system yecCS for an amino acid. Probably responsible for energy coupling to the transport system
- Gene Name:
- yecC
- Locus Tag:
- PA5152
- Molecular weight:
- 28.4 kDa
- General function:
- Involved in transporter activity
- Specific function:
- Part of the binding-protein-dependent transport system for histidine; probably responsible for the translocation of the substrate across the membrane
- Gene Name:
- hisM
- Locus Tag:
- PA2925
- Molecular weight:
- 26.7 kDa
- General function:
- Involved in transport
- Specific function:
- Permease that is involved in the transport across the cytoplasmic membrane of the aromatic amino acids (phenylalanine, tyrosine, and tryptophan)
- Gene Name:
- aroP
- Locus Tag:
- PA3000
- Molecular weight:
- 51 kDa
- General function:
- Involved in transporter activity
- Specific function:
- Part of the binding-protein-dependent transport system for histidine; probably responsible for the translocation of the substrate across the membrane
- Gene Name:
- hisQ
- Locus Tag:
- PA2924
- Molecular weight:
- 24.5 kDa
- General function:
- Involved in transporter activity
- Specific function:
- Component of the high-affinity histidine permease, a binding-protein-dependent transport system. The other components are proteins hisQ, hisM, and hisP
- Gene Name:
- hisJ
- Locus Tag:
- PA2923
- Molecular weight:
- 28.2 kDa