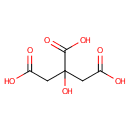
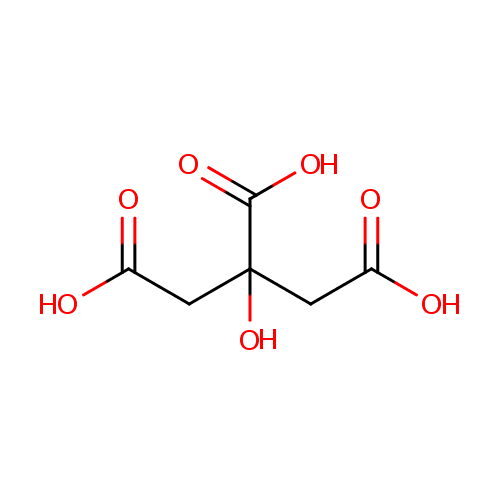
| References: |
- A J, Trygg J, Gullberg J, Johansson AI, Jonsson P, Antti H, Marklund SL, Moritz T: Extraction and GC/MS analysis of the human blood plasma metabolome. Anal Chem. 2005 Dec 15;77(24):8086-94. Pubmed: 16351159
- Abe T, Kobata H, Hanba Y, Kitabata Y, Narukawa N, Hasegawa H, Abe T, Fukagawa M: Study of plasma exchange for liver failure: beneficial and harmful effects. Ther Apher Dial. 2004 Jun;8(3):180-4. Pubmed: 15154867
- Bairaktari E, Katopodis K, Siamopoulos KC, Tsolas O: Paraquat-induced renal injury studied by 1H nuclear magnetic resonance spectroscopy of urine. Clin Chem. 1998 Jun;44(6 Pt 1):1256-61. Pubmed: 9625050
- Batchelor WB, Tolleson TR, Huang Y, Larsen RL, Mantell RM, Dillard P, Davidian M, Zhang D, Cantor WJ, Sketch MH Jr, Ohman EM, Zidar JP, Gretler D, DiBattiste PM, Tcheng JE, Califf RM, Harrington RA: Randomized COMparison of platelet inhibition with abciximab, tiRofiban and eptifibatide during percutaneous coronary intervention in acute coronary syndromes: the COMPARE trial. Comparison Of Measurements of Platelet aggregation with Aggrastat, Reopro, and Eptifibatide. Circulation. 2002 Sep 17;106(12):1470-6. Pubmed: 12234950
- Bennett, B. D., Kimball, E. H., Gao, M., Osterhout, R., Van Dien, S. J., Rabinowitz, J. D. (2009). "Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli." Nat Chem Biol 5:593-599. Pubmed: 19561621
- Caudarella R, Vescini F, Buffa A, Stefoni S: Citrate and mineral metabolism: kidney stones and bone disease. Front Biosci. 2003 Sep 1;8:s1084-106. Pubmed: 12957820
- Chiueh CC, Andoh T, Lai AR, Lai E, Krishna G: Neuroprotective strategies in Parkinson's disease: protection against progressive nigral damage induced by free radicals. Neurotox Res. 2000 May;2(2-3):293-310. Pubmed: 16787846
- Commodari F, Arnold DL, Sanctuary BC, Shoubridge EA: 1H NMR characterization of normal human cerebrospinal fluid and the detection of methylmalonic acid in a vitamin B12 deficient patient. NMR Biomed. 1991 Aug;4(4):192-200. Pubmed: 1931558
- Costello LC, Franklin RB: Testosterone and prolactin regulation of metabolic genes and citrate metabolism of prostate epithelial cells. Horm Metab Res. 2002 Aug;34(8):417-24. Pubmed: 12198595
- Hoffmann GF, Meier-Augenstein W, Stockler S, Surtees R, Rating D, Nyhan WL: Physiology and pathophysiology of organic acids in cerebrospinal fluid. J Inherit Metab Dis. 1993;16(4):648-69. Pubmed: 8412012
- Jurasovic J, Cvitkovic P, Pizent A, Colak B, Telisman S: Semen quality and reproductive endocrine function with regard to blood cadmium in Croatian male subjects. Biometals. 2004 Dec;17(6):735-43. Pubmed: 15689116
- Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
- Keseler, I. M., Collado-Vides, J., Santos-Zavaleta, A., Peralta-Gil, M., Gama-Castro, S., Muniz-Rascado, L., Bonavides-Martinez, C., Paley, S., Krummenacker, M., Altman, T., Kaipa, P., Spaulding, A., Pacheco, J., Latendresse, M., Fulcher, C., Sarker, M., Shearer, A. G., Mackie, A., Paulsen, I., Gunsalus, R. P., Karp, P. D. (2011). "EcoCyc: a comprehensive database of Escherichia coli biology." Nucleic Acids Res 39:D583-D590. Pubmed: 21097882
- Mairiang E, Hanpanich P, Sriboonlue P: In vivo 31P-MRS assessment of muscle-pH, cytolsolic-[Mg2+] and phosphorylation potential after supplementing hypokaliuric renal stone patients with potassium and magnesium salts. Magn Reson Imaging. 2004 Jun;22(5):715-9. Pubmed: 15172066
- McBane RD 2nd, Karnicki K, Miller RS, Owen WG: The impact of peripheral arterial disease on circulating platelets. Thromb Res. 2004;113(2):137-45. Pubmed: 15115669
- McBane RD 2nd, Karnicki K, Tahirkheli N, Miller RS, Owen WG: Platelet characteristics associated with coronary artery disease. J Thromb Haemost. 2003 Jun;1(6):1296-303. Pubmed: 12871333
- Moreno AJ, Brown JM, Salinas JA, Feaster BL 3rd, Brown TJ: Ga-67 positivity in sarcoidosis of the skin with coincident thyroid uptake of uncertain etiology. Clin Nucl Med. 1984 Mar;9(3):165-6. Pubmed: 6705419
- Odvina CV, Mason RP, Pak CY: Prevention of thiazide-induced hypokalemia without magnesium depletion by potassium-magnesium-citrate. Am J Ther. 2006 Mar-Apr;13(2):101-8. Pubmed: 16645424
- Seiffert D, Thomas BE, Bradley JD, Munzer DA, Tchinnes MA, Kornhauser DM, Cain VA, Hua TA, Feuerstein GZ, Martin DE, Stern AM: Effects of the glycoprotein IIb/IIIa antagonist Roxifiban on P-selectin expression, fibrinogen binding, and microaggregate formation in a phase I dose-finding study: no evidence for platelet activation during treatment with a glycoprotein IIb/IIIa antagonist. Platelets. 2003 May;14(3):179-87. Pubmed: 12850842
- Solheim BG, Flesland O, Brosstad F, Mollnes TE, Seghatchian J: Improved preservation of coagulation factors after pre-storage leukocyte depletion of whole blood. Transfus Apher Sci. 2003 Oct;29(2):133-9. Pubmed: 12941351
- Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, Nyati MK, Ahsan A, Kalyana-Sundaram S, Han B, Cao X, Byun J, Omenn GS, Ghosh D, Pennathur S, Alexander DC, Berger A, Shuster JR, Wei JT, Varambally S, Beecher C, Chinnaiyan AM: Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009 Feb 12;457(7231):910-4. Pubmed: 19212411
- Tammen H, Schulte I, Hess R, Menzel C, Kellmann M, Mohring T, Schulz-Knappe P: Peptidomic analysis of human blood specimens: comparison between plasma specimens and serum by differential peptide display. Proteomics. 2005 Aug;5(13):3414-22. Pubmed: 16038021
- van der Werf, M. J., Overkamp, K. M., Muilwijk, B., Coulier, L., Hankemeier, T. (2007). "Microbial metabolomics: toward a platform with full metabolome coverage." Anal Biochem 370:17-25. Pubmed: 17765195
- Vijayendran, C., Barsch, A., Friehs, K., Niehaus, K., Becker, A., Flaschel, E. (2008). "Perceiving molecular evolution processes in Escherichia coli by comprehensive metabolite and gene expression profiling." Genome Biol 9:R72. Pubmed: 18402659
- Wan KW, Malgesini B, Verpilio I, Ferruti P, Griffiths PC, Paul A, Hann AC, Duncan R: Poly(amidoamine) salt form: effect on pH-dependent membrane activity and polymer conformation in solution. Biomacromolecules. 2004 May-Jun;5(3):1102-9. Pubmed: 15132705
- Wei R, Han JJ, Bai B, Ren DL, Chen B, Yang MF, Xia ZL: Analysis of factors influencing the blood levels and activities of tissue-type plasminogen activator (t-PA). Clin Hemorheol Microcirc. 2003;29(3-4):351-6. Pubmed: 14724361
- Wenk J, Foitzik A, Achterberg V, Sabiwalsky A, Dissemond J, Meewes C, Reitz A, Brenneisen P, Wlaschek M, Meyer-Ingold W, Scharffetter-Kochanek K: Selective pick-up of increased iron by deferoxamine-coupled cellulose abrogates the iron-driven induction of matrix-degrading metalloproteinase 1 and lipid peroxidation in human dermal fibroblasts in vitro: a new dressing concept. J Invest Dermatol. 2001 Jun;116(6):833-9. Pubmed: 11407968
- Wevers RA, Engelke U, Wendel U, de Jong JG, Gabreels FJ, Heerschap A: Standardized method for high-resolution 1H-NMR of cerebrospinal fluid. Clin Chem. 1995 May;41(5):744-51. Pubmed: 7729054
- Winder, C. L., Dunn, W. B., Schuler, S., Broadhurst, D., Jarvis, R., Stephens, G. M., Goodacre, R. (2008). "Global metabolic profiling of Escherichia coli cultures: an evaluation of methods for quenching and extraction of intracellular metabolites." Anal Chem 80:2939-2948. Pubmed: 18331064
|
|---|