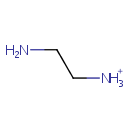
Search Results for compounds
Searching compounds for
returned 4373 results.
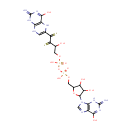
N-Acetylmuramoyl-L-alanyl-D-glutamyl-meso-2,6-diaminopimelyl-D-alanyl-D-alanine-diphosphoundecaprenol (PAMDB001100)
IUPAC:
(2R,6S)-2-amino-6-{[(4R)-4-carboxy-1-hydroxy-4-{[(2S)-1-hydroxy-2-{[(2R)-1-hydroxy-2-{[(2R,3S,4R,5R,6S)-3-hydroxy-6-{[hydroxy({hydroxy[(3,7,11,15,19,23,27,31,35,39,43-undecamethyltetratetraconta-2,6,10,14,18,22,26,30,34,38,42-undecaen-1-yl)oxy]phosphoryl}oxy)phosphoryl]oxy}-5-[(1-hydroxyethylidene)amino]-2-(hydroxymethyl)oxan-4-yl]oxy}propylidene]amino}propylidene]amino}butylidene]amino}-6-{[(1R)-1-{[(1R)-1-carboxyethyl]-C-hydroxycarbonimidoyl}ethyl]-C-hydroxycarbonimidoyl}hexanoic acid
CAS: Not Available
Description: N-acetylmuramoyl-L-alanyl-D-glutamyl-meso-2,6-diaminopimelyl-D-alanyl-D-alanine-diphosphoundecaprenol is an intermediate in peptidoglycan synthesis. It is a substrate for the enzyme undecaprenyldiphospho-muramoylpentapeptide beta-N-acetylglucosaminyltransferase (murG). Peptidoglycan can be described as a fisherman's net that encloses bacteria. The mesh of the net is made of two segments of parallel, somewhat inextensible glycan threads, held together by two small elastic peptide crosslinks allowing the net to expand or shrink. The glycan moiety of the peptidoglycan is very uniform among all bacteria, and is made up of alternating β-1,4-linked N-acetylglucosamine and N-acetyl muramate residues, with an average chain lengthof 10 to 65 disaccharide units (depending on the organism). The peptidoglycan synthesis pathway starts in the cytoplasm, where in six steps the peptidoglycan precursor a UDP-N-acetylmuramoyl-pentapeptide is synthesized. This precursor is then attached to the memberane acceptor all-trans-undecaprenyl phosphate, generating a N-acetylmuramoyl-pentapeptide-diphosphoundecaprenol, also known as lipid I. Another transferase then adds UDP-N-acetyl-α-D-glucosamine, yielding the complete monomeric unit a lipid II, also known as lipid II. This final lipid intermediate is transferred by an as yet unknown mechanism through the membrane. The peptidoglycan monomers are then polymerized on the outside surface by glycosyltransferases, which form the linear glycan chains, and transpeptidases, which catalyze the formation of peptide crosslinks. Peptide crosslinks form between different parts of the peptides depending on the organism. For example, in Mycobacteria and in Pseudomonas aeruginosa most links form between the carboxyl group of the penultimate D-alanine (residue 4) of one peptide to the amino group at the D-center of meso-diaminopimelate (residue 3) of an adjacent peptide of a second glycan chain (as in Pseudomonas aeruginosa). The crosslinking reaction is catalyzed by transpeptidases and involves the cleavage of the D-alanyl-D-alanine bond of the donor peptide, providing the energy to drive the reaction. As a result, the peptides in the peptidoglycan polymers are one or two amino acids shorter than the peptides in the monomers.Glycosyltransferase MurG catalyses the transfer of N-acetyl-d-glucosamine to lipid intermediate I on the bacterial peptidoglycan biosynthesis pathway, and is a target for development of new antibacterial agents. (PMID 20226679)
Ethylenediamine (PAMDB001114)
IUPAC:
2-aminoethan-1-aminium
CAS: 333-18-6
Description: Ethylenediamine (abbreviated as en when a ligand) is the organic compound with the formula C2H4(NH2)2. This colorless liquid with an ammonia-like odor is a strongly basic amine. It is a widely used building block in chemical synthesis, with approximately 500,000,000 kg being produced in 1998.In terms of quantities produced, ethylenediamine is the most significant diamine (aside from diaminohexane, which is a precursor to Nylon 6-6). Related derivatives of ethylenediamine include tetramethylethylenediamine, abbreviated (TMEDA), (CH3)2N-CH2CH2-N(CH3)2 and tetraethylethylenediamine, abbreviated (TEEDA), (C2H5)2N-CH2CH2-N(C2H5)2The bleaching activator tetraacetylethylenediamine is generated from ethylenediamine. The derivative N,N-ethylenebis(stearamide) (EBS) is a commercially significant mold-release agent and a surfactant in gasoline and motor oil."
Molybdopterin guanine dinucleotide (PAMDB001121)
IUPAC:
[({[3,4-dihydroxy-5-(6-hydroxy-2-imino-3,9-dihydro-2H-purin-9-yl)oxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy][2-hydroxy-4-(4-hydroxy-2-imino-1,2,5,8-tetrahydropteridin-6-yl)-3,4-disulfanylidenebutoxy]phosphinic acid
CAS: 128007-95-4
Description: Molybdopterin guanine dinucleotide is a member of the chemical class known as Purine Ribonucleoside Diphosphates. These are purine ribobucleotides with diphosphate group linked to the ribose moiety. It appears that the molybdopterin present in the nitrate reductase of a chlB mutant is converted to molybdopterin guanine dinucleotide during activation. (PMID 1459941) We propose therefore that MobB is an adapter protein that acts in concert with MobA to achieve the efficient biosynthesis and utilization of molybdopterin guanine dinucleotide. (PMID 12682065)
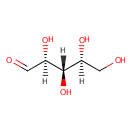
L-Ribose (PAMDB001122)
IUPAC:
(2S,3S,4S)-2,3,4,5-tetrahydroxypentanal
CAS: 24259-59-4
Description: Ribose is an organic compound that occurs widely in nature. It is an aldopentose, a monosaccharide containing five carbon atoms that in its acyclic form has an aldehyde functional group at one end. Typically, ribose exists in the cyclic form. It comprises the backbone of RNA, a biopolymer that is the basis of genetic transcription. It is related to deoxyribose, as found in DNA, by the removal of one hydroxy group. Once phosphorylated, ribose can become a subunit of ATP, NADH, and several other compounds that are critical to metabolism. Ribose exists in two enantiomeric forms, primarily as D-ribose. L-ribose is the synthetic mirror image of D-ribose. (Wikipedia) In Pseudomonas aeruginosa, L-ribose can act as an competitive inhibitor of glucose dehydrogenase. (EcoCyc)
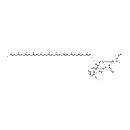
N-Acetylmuramoyl-L-alanyl-D-glutamyl-L-lysyl-D-alanyl-D-alanine-diphosphoundecaprenyl-N-acetylglucosamine (PAMDB001129)
IUPAC:
(4R)-4-{[(1S)-5-amino-1-{[(1R)-1-{[(1R)-1-carboxyethyl]-C-hydroxycarbonimidoyl}ethyl]-C-hydroxycarbonimidoyl}pentyl]-C-hydroxycarbonimidoyl}-4-{[(2S)-2-{[(2R)-2-{[(2R,3S,4R,5R,6R)-3-{[(2R,3R,4R,5S,6R)-4,5-dihydroxy-3-[(1-hydroxyethylidene)amino]-6-(hydroxymethyl)oxan-2-yl]oxy}-6-{[hydroxy({hydroxy[(3,7,11,15,19,23,27,31,35,39,43-undecamethyltetratetraconta-2,6,10,14,18,22,26,30,34,38,42-undecaen-1-yl)oxy]phosphoryl}oxy)phosphoryl]oxy}-5-[(1-hydroxyethylidene)amino]-2-(hydroxymethyl)oxan-4-yl]oxy}-1-hydroxypropylidene]amino}-1-hydroxypropylidene]amino}butanoic acid
CAS: Not Available
Description: N-acetylmuramoyl-L-alanyl-D-glutamyl-L-lysyl-D-alanyl-D-alanine-diphosphoundecaprenyl-N-acetylglucosamine is an intermediate in peptidoglycan synthesis. It is a substrate for the enzyme undecaprenyldiphospho-muramoylpentapeptide beta-N-acetylglucosaminyltransferase (murG). Peptidoglycan can be described as a fisherman's net that encloses bacteria. The mesh of the net is made of two segments of parallel, somewhat inextensible glycan threads, held together by two small elastic peptide crosslinks allowing the net to expand or shrink. The glycan moiety of the peptidoglycan is very uniform among all bacteria, and is made up of alternating β-1,4-linked N-acetylglucosamine and N-acetyl muramate residues, with an average chain lengthof 10 to 65 disaccharide units (depending on the organism). The peptidoglycan synthesis pathway starts in the cytoplasm, where in six steps the peptidoglycan precursor a UDP-N-acetylmuramoyl-pentapeptide is synthesized. This precursor is then attached to the memberane acceptor all-trans-undecaprenyl phosphate, generating a N-acetylmuramoyl-pentapeptide-diphosphoundecaprenol, also known as lipid I. Another transferase then adds UDP-N-acetyl-α-D-glucosamine, yielding the complete monomeric unit a lipid II, also known as lipid II. This final lipid intermediate is transferred by an as yet unknown mechanism through the membrane. The peptidoglycan monomers are then polymerized on the outside surface by glycosyltransferases, which form the linear glycan chains, and transpeptidases, which catalyze the formation of peptide crosslinks. Peptide crosslinks form between different parts of the peptides depending on the organism. For example, in Mycobacteria and in Pseudomonas aeruginosa most links form between the carboxyl group of the penultimate D-alanine (residue 4) of one peptide to the amino group at the D-center of meso-diaminopimelate (residue 3) of an adjacent peptide of a second glycan chain (as in Pseudomonas aeruginosa). The crosslinking reaction is catalyzed by transpeptidases and involves the cleavage of the D-alanyl-D-alanine bond of the donor peptide, providing the energy to drive the reaction. As a result, the peptides in the peptidoglycan polymers are one or two amino acids shorter than the peptides in the monomers.
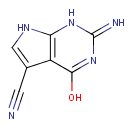
7-Cyano-7-carbaguanine (PAMDB001132)
IUPAC:
4-hydroxy-2-imino-1H,2H,7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile
CAS: Not Available
Description: 7-cyano-7-carbaguanine is a member of the chemical class known as Pyrrolopyrimidines. These are compounds containing a pyrrolopyrimidine moiety, which consists of a pyrrole ring fused to a pyrimidine. This compound is involved in queuosine biosynthesis.
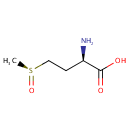
L-Methionine-(S)-S-oxide (PAMDB001133)
IUPAC:
(2R)-2-amino-4-[(S)-methanesulfinyl]butanoic acid
CAS: 454-41-1
Description: L-methionine-(s)-s-oxide is an oxidized form of methionine. Methionine is an amino acid susceptible to being oxidized to methionine sulfoxide (MetSO). The reduction of MetSO to methionine is catalyzed by methionine sulfoxide reductase (MSR), an enzyme present in almost all organisms. (PMID 20969952) Oxidation of methionine to methionine sulfoxide is a major oxidative stress product that reaches levels as high as 60% in cataract while being essentially absent from clear lenses. Methionine oxidation results in loss of protein function that can be reversed through the action of methionine sulfoxide reductase A (MsrA), which is implicated in oxidative stress protection and is an essential regulator of longevity in species ranging from Pseudomonas aeruginosa to mice. (PMID 15199188)
TDP-Glucose (PAMDB001135)
IUPAC:
1-[(2R,4S,5R)-4-hydroxy-5-({[hydroxy({[(2R,3R,4S,5S,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy})phosphoryl phosphonato]oxy}methyl)oxolan-2-yl]-5-methyl-2-oxo-1,2-dihydropyrimidin-4-olate
CAS: Not Available
Description: TDP-glucose is a member of the chemical class known as Pyrimidine Nucleotide Sugars. These are pyrimidine nucleotides bound to a saccharide derivative through the terminal phosphate group. Thymidine diphosphate glucose (often abbreviated dTDP-glucose or TDP-glucose) is a nucleotide-linked sugar consisting of deoxythymidine diphosphate linked to glucose. It is the starting compound for the syntheses of many deoxysugars. (WikiPedia)
Di-trans,poly-cis-undecaprenyl phosphate (PAMDB001145)
IUPAC:
{dioxo[(3,7,11,15,19,23,27,31,35,39,43-undecamethyltetratetraconta-2,6,10,14,18,22,26,30,34,38,42-undecaen-1-yl)oxy]-???phosphanuidyl}oxidanyl
CAS: Not Available
Description: Di-trans,poly-cis-undecaprenyl phosphate is an undecaprenyl phosphate having two (E)- and eight (Z)-double bonds. It is invovled in Peptidoglycan biosynthesis. (KEGG). Pseudomonas aeruginosa and other Gram-negative bacteria utilize the methylerythritol phosphate pathway to synthesize isopentenyl diphosphate and dimethylallyl diphosphate which are the precursors of (2E,6E)-farnesyl diphosphate. The latter compound is a precursor of di-trans,poly-cis-undecaprenyl phosphate, which leads to the biosynthesis of cell wall polymers. Initially (2E,6E)-farnesyl diphosphate is formed by the joining of geranyl diphosphate and isopentenyl diphosphate in a reaction catalyzed by geranyl diphosphate synthase / farnesyl diphosphate synthase encoded by gene ispA. Subsequently multiple units of isopentenyl diphosphate are polymerized to form di-trans,octa-cis-undecaprenyl diphosphate in a series of sequential condensation reactions catalyzed by undecaprenyl diphosphate synthase encoded by gene ispU.
Decarboxy-SAM (PAMDB001159)
IUPAC:
{[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl}(3-azaniumylpropyl)methylsulfanium
CAS: Not Available
Description: dcSAM is a member of the chemical class known as Purine Nucleosides and Analogues. These are compounds comprising a purine base attached to a sugar. [2-(Amino-oxy)ethyl](5'-deoxyadenosin-5'-yl)(methyl)sulphonium+ ++, the amino-oxy analogue of decarboxylated S-adenosylmethionine, is a potent irreversible inhibitor of Pseudomonas aeruginosa S-adenosylmethionine decarboxylase [Khomutov, Zavalova, Syrku, Artamonova & Khomutov (1983) Bioorg. (PMID 1872800)