Search Results for compounds
Searching compounds for
returned 4373 results.
Displaying compounds 1091 - 1100 of
4373 in total
Retinal (PAMDB002049)
IUPAC:
(2E,4E,6E,8E)-3,7-dimethyl-9-(2,6,6-trimethylcyclohex-1-en-1-yl)nona-2,4,6,8-tetraenal
CAS: 116-31-4
Description: A carotenoid constituent of visual pigments. It is the oxidized form of retinol which functions as the active component of the visual cycle. It is bound to the protein opsin forming the complex rhodopsin. When stimulated by visible light, the retinal component of the rhodopsin complex undergoes isomerization at the 11-position of the double bond to the cis-form; this is reversed in 'dark' reactions to return to the native trans-configuration.; Retinal, also called retinaldehyde or vitamin A aldehyde, is one of the many forms of vitamin A (the number of which varies from species to species). Retinal is a polyene chromophore, and bound to proteins called opsins, is the chemical basis of animal vision. Bound to proteins called type 1 rhodopsins, retinal allows certain microorganisms to convert light into metabolic energy.; The visual cycle is a circular enzymatic pathway, which is the front-end of phototransduction. It regenerates 11-cis-retinal.; Vertebrate animals ingest retinal directly from meat, or produce retinal from one of four carotenoids (beta-carotene, alpha-carotene, gamma-carotene, and beta-cryptoxanthin), which they must obtain from plants or other photosynthetic organisms (no other carotenoids can be converted by animals to retinal, and some carnivores cannot convert any carotenoids at all). The other main forms of vitamin A, retinol, and a partially active form retinoic acid, may both be produced from retinal.; catalyzed by retinal dehydrogenases also known as retinaldehyde dehydrogenases (RALDHs) as well as retinal oxidases. Retinoic acid, sometimes called vitamin A acid, is an important signaling molecule and hormone in vertebrate animals.
Retinol (PAMDB002050)
IUPAC:
(2E,4E,6E,8E)-3,7-dimethyl-9-(2,6,6-trimethylcyclohex-1-en-1-yl)nona-2,4,6,8-tetraen-1-ol
CAS: 68-26-8
Description: Retinol is also known as Vitamin A. In Pseudomonas aeruginosa, retinol can be oxidized to retinal in a reversible reaction catalyzed by alcohol dehydrogenases (EC 1.1.1.1) such as those encoded by Pseudomonas aeruginosa genes frmA and adhP. This reaction also reduces NAD+ to NADH. (KEGG)
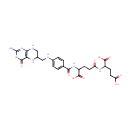
Tetrahydrofolyl-[Glu](2) (PAMDB002051)
IUPAC:
2-{4-[(4-{[(2-amino-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl)methyl]amino}phenyl)formamido]-4-carboxybutanamido}pentanedioic acid
CAS: Not Available
Description: Tetrahydrofolyl-[Glu](n) is involved in the folate biosynthesis pathway. Tetrahydrofolyl-[Glu](n) can be reversibly converted into Tetrahydrofolyl-[Glu](2) by folylpolyglutamate synthase [EC:6.3.2.17]. Tetrahydrofolyl-[Glu](n) can be irreversibly converted into tetrahydrofolate by gamma-glutamyl hydrolase [EC:3.4.19.9].
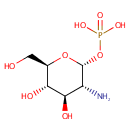
alpha-D-Glucosamine 1-phosphate (PAMDB002052)
IUPAC:
{[(2R,3R,4R,5S,6R)-3-amino-4,5-dihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}phosphonic acid
CAS: Not Available
Description: Alpha-D-glucosamine 1-phosphate is a member of the chemical class known as Hexoses. These are monosaccharides in which the sugar unit is a hexose. It is involved and amino sugar and nucleotide sugar metabolism.
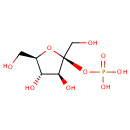
beta-D-Fructose 2-phosphate (PAMDB002053)
IUPAC:
{[(2S,3S,4S,5R)-3,4-dihydroxy-2,5-bis(hydroxymethyl)oxolan-2-yl]oxy}phosphonic acid
CAS: 19046-69-6
Description: Beta-D-Fructose 2-phosphate is involved in the fructose eand mannose system. beta-D-Fructose 2-phosphate is produced from beta-D-Fructose 2,6-bisphosphate by the enzyme fructose-2,6-bisphosphate 6-phosphatase [EC 3.1.3.54].
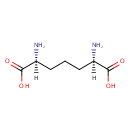
Meso-2,6-Diaminoheptanedioate (PAMDB002055)
IUPAC:
(2R,6S)-2,6-diaminoheptanedioic acid
CAS: 922-54-3
Description: Meso-2,6-diaminoheptanedioate is a member of the chemical class known as Alpha Amino Acids and Derivatives. These are amino acids in which the amino group is attached to the carbon atom immediately adjacent to the carboxylate group (alpha carbon). This particular compound is at a branch point in lysine biosynthesis where it is either the penultimate step in lysine biosynthesis in Pseudomonas aeruginosa (via diaminopimelate decarboxylase, LysA) or it can be directed towards the synthesis of peptidoglycan (via murE).
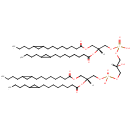
CL(15:0cyclo/15:0cyclo/15:0cyclo/15:0cyclo) (PAMDB002056)
IUPAC:
[(2R)-2,3-bis({[8-(2-butylcyclopropyl)octanoyl]oxy})propoxy][3-({[(2R)-2,3-bis({[8-(2-butylcyclopropyl)octanoyl]oxy})propoxy](hydroxy)phosphoryl}oxy)-2-hydroxypropoxy]phosphinic acid
CAS: Not Available
Description: CL(15:0cyclo/15:0cyclo/15:0cyclo/15:0cyclo) is a cardiolipin (CL). Cardiolipins are sometimes called a 'double' phospholipid because they have four fatty acid tails, instead of the usual two. CL(15:0cyclo/15:0cyclo/15:0cyclo/15:0cyclo) contains four chains of cis-9,10-Methylenetetradecanoic acid at the C1, C2, C3 and C4 positions. While the theoretical charge of cardiolipins is -2, under normal physiological conditions (pH near 7), the molecule may carry only one negative charge. In prokaryotes such as Pseudomonas aeruginosa, the enzyme known as diphosphatidylglycerol synthase catalyses the transfer of the phosphatidyl moiety of one phosphatidylglycerol to the free 3'-hydroxyl group of another, with the elimination of one molecule of glycerol. In Pseudomonas aeruginosa, which acylates its glycerophospholipids with acyl chains ranging in length from 12 to 19 carbons and possibly containing an unsaturation, or a cyclopropane group more than 100 possible CL molecular species are theoretically possible. Pseudomonas aeruginosa membranes consist of ~5% cardiolipin (CL), 20-25% phosphatidylglycerol (PG), and 70-80% phosphatidylethanolamine (PE) as well as smaller amounts of phosphatidylserine (PS). CL is distributed between the two leaflets of the bilayers and is located preferentially at the poles and septa in Pseudomonas aeruginosa and other rod-shaped bacteria. It is known that the polar positioning of the proline transporter ProP and the mechanosensitive ion channel MscS in Pseudomonas aeruginosa is dependent on CL. It is believed that cell shape may influence the localization of CL and the localization of certain membrane proteins.
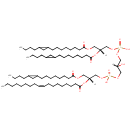
CL(15:0cyclo/15:0cyclo/15:0cyclo/18:1(9Z)) (PAMDB002059)
IUPAC:
[(2R)-2,3-bis({[8-(2-butylcyclopropyl)octanoyl]oxy})propoxy][(2R)-3-({[(2R)-3-{[8-(2-butylcyclopropyl)octanoyl]oxy}-2-[(9Z)-octadec-9-enoyloxy]propoxy](hydroxy)phosphoryl}oxy)-2-hydroxypropoxy]phosphinic acid
CAS: Not Available
Description: CL(15:0cyclo/15:0cyclo/15:0cyclo/18:1(9Z)) is a cardiolipin (CL). Cardiolipins are sometimes called a 'double' phospholipid because they have four fatty acid tails, instead of the usual two. CL(15:0cyclo/15:0cyclo/15:0cyclo/18:1(9Z)) contains three chains of cis-9,10-Methylenetetradecanoic acid at the C1, C2 and C3 positions, one chain of (9Z-octadecenoyl) at the C4 position. While the theoretical charge of cardiolipins is -2, under normal physiological conditions (pH near 7), the molecule may carry only one negative charge. In prokaryotes such as Pseudomonas aeruginosa, the enzyme known as diphosphatidylglycerol synthase catalyses the transfer of the phosphatidyl moiety of one phosphatidylglycerol to the free 3'-hydroxyl group of another, with the elimination of one molecule of glycerol. In Pseudomonas aeruginosa, which acylates its glycerophospholipids with acyl chains ranging in length from 12 to 19 carbons and possibly containing an unsaturation, or a cyclopropane group more than 100 possible CL molecular species are theoretically possible. Pseudomonas aeruginosa membranes consist of ~5% cardiolipin (CL), 20-25% phosphatidylglycerol (PG), and 70-80% phosphatidylethanolamine (PE) as well as smaller amounts of phosphatidylserine (PS). CL is distributed between the two leaflets of the bilayers and is located preferentially at the poles and septa in Pseudomonas aeruginosa and other rod-shaped bacteria. It is known that the polar positioning of the proline transporter ProP and the mechanosensitive ion channel MscS in Pseudomonas aeruginosa is dependent on CL. It is believed that cell shape may influence the localization of CL and the localization of certain membrane proteins.
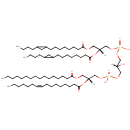
CL(15:0cyclo/15:0cyclo/15:0cyclo/19:0cycv8c) (PAMDB002060)
IUPAC:
[(2R)-3-({[(2R)-2,3-bis({[8-(2-butylcyclopropyl)octanoyl]oxy})propoxy](hydroxy)phosphoryl}oxy)-2-hydroxypropoxy][(2R)-3-{[8-(2-butylcyclopropyl)octanoyl]oxy}-2-{[10-(2-hexylcyclopropyl)decanoyl]oxy}propoxy]phosphinic acid
CAS: Not Available
Description: CL(15:0cyclo/15:0cyclo/15:0cyclo/19:0cycv8c) is a cardiolipin (CL). Cardiolipins are sometimes called a 'double' phospholipid because they have four fatty acid tails, instead of the usual two. CL(15:0cyclo/15:0cyclo/15:0cyclo/19:0cycv8c) contains three chains of cis-9,10-Methylenetetradecanoic acid at the C1, C2 and C3 positions, one chain of (heptadec-11-12-cyclo-anoyl) at the C4 position. While the theoretical charge of cardiolipins is -2, under normal physiological conditions (pH near 7), the molecule may carry only one negative charge. In prokaryotes such as Pseudomonas aeruginosa, the enzyme known as diphosphatidylglycerol synthase catalyses the transfer of the phosphatidyl moiety of one phosphatidylglycerol to the free 3'-hydroxyl group of another, with the elimination of one molecule of glycerol. In Pseudomonas aeruginosa, which acylates its glycerophospholipids with acyl chains ranging in length from 12 to 19 carbons and possibly containing an unsaturation, or a cyclopropane group more than 100 possible CL molecular species are theoretically possible. Pseudomonas aeruginosa membranes consist of ~5% cardiolipin (CL), 20-25% phosphatidylglycerol (PG), and 70-80% phosphatidylethanolamine (PE) as well as smaller amounts of phosphatidylserine (PS). CL is distributed between the two leaflets of the bilayers and is located preferentially at the poles and septa in Pseudomonas aeruginosa and other rod-shaped bacteria. It is known that the polar positioning of the proline transporter ProP and the mechanosensitive ion channel MscS in Pseudomonas aeruginosa is dependent on CL. It is believed that cell shape may influence the localization of CL and the localization of certain membrane proteins.
CL(15:0cyclo/15:0cyclo/16:0/16:1(9Z)) (PAMDB002065)
IUPAC:
[(2R)-2,3-bis({[8-(2-butylcyclopropyl)octanoyl]oxy})propoxy][(2R)-3-({[(2R)-2-[(9Z)-hexadec-9-enoyloxy]-3-(hexadecanoyloxy)propoxy](hydroxy)phosphoryl}oxy)-2-hydroxypropoxy]phosphinic acid
CAS: Not Available
Description: CL(15:0cyclo/15:0cyclo/16:0/16:1(9Z)) is a cardiolipin (CL). Cardiolipins are sometimes called a 'double' phospholipid because they have four fatty acid tails, instead of the usual two. CL(15:0cyclo/15:0cyclo/16:0/16:1(9Z)) contains two chains of cis-9,10-Methylenetetradecanoic acid at the C1 and C2 positions, one chain of hexadecanoic acid at the C3 position, one chain of (9Z-hexadecenoyl) at the C4 position. While the theoretical charge of cardiolipins is -2, under normal physiological conditions (pH near 7), the molecule may carry only one negative charge. In prokaryotes such as Pseudomonas aeruginosa, the enzyme known as diphosphatidylglycerol synthase catalyses the transfer of the phosphatidyl moiety of one phosphatidylglycerol to the free 3'-hydroxyl group of another, with the elimination of one molecule of glycerol. In Pseudomonas aeruginosa, which acylates its glycerophospholipids with acyl chains ranging in length from 12 to 19 carbons and possibly containing an unsaturation, or a cyclopropane group more than 100 possible CL molecular species are theoretically possible. Pseudomonas aeruginosa membranes consist of ~5% cardiolipin (CL), 20-25% phosphatidylglycerol (PG), and 70-80% phosphatidylethanolamine (PE) as well as smaller amounts of phosphatidylserine (PS). CL is distributed between the two leaflets of the bilayers and is located preferentially at the poles and septa in Pseudomonas aeruginosa and other rod-shaped bacteria. It is known that the polar positioning of the proline transporter ProP and the mechanosensitive ion channel MscS in Pseudomonas aeruginosa is dependent on CL. It is believed that cell shape may influence the localization of CL and the localization of certain membrane proteins.