| Identification |
| Name: |
UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase |
| Synonyms: |
- Protein envA
- UDP-3-O-acyl-GlcNAc deacetylase
|
| Gene Name: |
lpxC |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase activity |
| Specific Function: |
The key enzyme in the biosynthesis of lipid A, a phosphorylated glycolipid that anchors the lipopolysaccharide to the outer membrane of the cell. Degraded by FtsH; when the activity of FtsH is reduced too much lipid A and not enough phospholipids are made (both pathways use the same precursor), which is lethal |
| Cellular Location: |
Cytoplasmic |
| KEGG Pathways: |
|
| KEGG Reactions: |
|
| SMPDB Reactions: |
|
| PseudoCyc/BioCyc Reactions: |
|
| Complex Reactions: |
Not Available |
| Transports: |
Not Available |
| Metabolites: |
| PAMDB ID | Name | View |
|---|
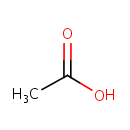
| PAMDB000012 | Acetic acid | MetaboCard | | PAMDB000644 | UDP-3-O-(3-Hydroxymyristoyl)-N-acetylglucosamine | MetaboCard | | PAMDB001696 | UDP-3-O-(3-Hydroxytetradecanoyl)-D-glucosamine | MetaboCard | | PAMDB006332 | UDP-3-O-[(3R)-3-hydroxymyristoyl]-N-acetyl-α-D-glucosamine | MetaboCard | | PAMDB000142 | Water | MetaboCard |
|
| GO Classification: |
| Function |
|---|
| catalytic activity | | hydrolase activity | | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds | | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides | | UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase activity | | Process |
|---|
| lipid A biosynthetic process | | metabolic process | | organophosphate metabolic process | | phospholipid biosynthetic process | | phospholipid metabolic process |
|
| Gene Properties |
| Locus tag: |
PA4406 |
| Strand: |
- |
| Entrez Gene ID: |
881292 |
| Accession: |
NP_253096.1 |
| GI: |
15599602 |
| Sequence start: |
4938276 |
| Sequence End: |
4939187 |
| Sequence Length: |
911 |
| Gene Sequence: |
>PA4406
ATGATCAAACAACGCACCTTGAAGAACATCATCCGGGCTACTGGCGTCGGTCTGCACTCGGGGGAAAAGGTTTACCTGACCCTGAAACCGGCGCCGGTGGACACCGGTATCGTGTTCTGCCGCACCGACCTGGATCCGGTCGTGGAAATCCCGGCCCGGGCCGAGAACGTCGGCGAAACCACCATGTCGACCACCCTGGTCAAGGGTGACGTCAAAGTGGATACGGTGGAGCACCTGCTCTCGGCCATGGCCGGCCTGGGTATCGACAACGCCTACGTCGAGCTGTCCGCTTCGGAAGTGCCGATCATGGATGGCAGCGCTGGTCCGTTCGTATTCCTGATCCAGTCCGCCGGATTGCAAGAGCAGGAAGCTGCCAAGAAGTTCATCCGCATCAAGCGCGAAGTCAGCGTGGAAGAGGGCGACAAGCGCGCCGTCTTCGTTCCGTTCGACGGCTTCAAGGTCAGCTTCGAGATCGATTTCGATCATCCGGTCTTCCGTGGTCGCACCCAGCAGGCCTCGGTCGATTTCTCCAGTACTTCCTTCGTCAAGGAGGTCAGCCGCGCCCGTACCTTCGGGTTCATGCGCGACATCGAGTACCTGCGTTCGCAGAACCTGGCACTCGGCGGTAGCGTGGAGAACGCGATCGTGGTCGACGAGAACCGCGTGCTCAACGAAGACGGCCTGCGTTACGAGGACGAGTTCGTCAAGCACAAGATCCTGGATGCCATCGGCGACCTGTATCTGCTCGGCAACAGTCTTATCGGCGAGTTCCGTGGCTTCAAGTCCGGCCATGCCCTGAACAACCAACTGCTGCGTACGTTGATCGCAGACAAGGATGCTTGGGAAGTGGTGACCTTCGAAGACGCGCGTACCGCGCCTATTTCCTATATGCGCCCGGCGGCGGCAGTGTAG |
| Protein Properties |
| Protein Residues: |
303 |
| Protein Molecular Weight: |
33.4 kDa |
| Protein Theoretical pI: |
5 |
| Hydropathicity (GRAVY score): |
-0.066 |
| Charge at pH 7 (predicted): |
-7.82 |
| Protein Sequence: |
>PA4406
MIKQRTLKNIIRATGVGLHSGEKVYLTLKPAPVDTGIVFCRTDLDPVVEIPARAENVGETTMSTTLVKGDVKVDTVEHLLSAMAGLGIDNAYVELSASEVPIMDGSAGPFVFLIQSAGLQEQEAAKKFIRIKREVSVEEGDKRAVFVPFDGFKVSFEIDFDHPVFRGRTQQASVDFSSTSFVKEVSRARTFGFMRDIEYLRSQNLALGGSVENAIVVDENRVLNEDGLRYEDEFVKHKILDAIGDLYLLGNSLIGEFRGFKSGHALNNQLLRTLIADKDAWEVVTFEDARTAPISYMRPAAAV |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA4406 and its homologs
|