| Identification |
| Name: |
Disulfide bond formation protein B |
| Synonyms: |
|
| Gene Name: |
dsbB |
| Enzyme Class: |
Not Available |
| Biological Properties |
| General Function: |
Involved in protein disulfide oxidoreductase activity |
| Specific Function: |
Required for disulfide bond formation in some periplasmic proteins such as phoA or ompA. Acts by oxidizing the dsbA protein |
| Cellular Location: |
Cell inner membrane; Multi-pass membrane protein |
| KEGG Pathways: |
|
| KEGG Reactions: |
Not Available |
| SMPDB Reactions: |
Not Available |
| PseudoCyc/BioCyc Reactions: |
|
Protein-Red-Disulfides | ↔ | Protein-Ox-Disulfides |
| |
|
| Complex Reactions: |
|
 | + | periplasmic protein disulfide isomerase I (reduced) | → |  | + | periplasmic protein disulfide isomerase I (oxidized) |
| | |
Menaquinone 8 | + | periplasmic protein disulfide isomerase I (reduced) | → | 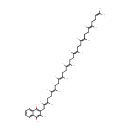 | + | periplasmic protein disulfide isomerase I (oxidized) |
| Menaquinone 8 + periplasmic protein disulfide isomerase I (reduced) → Menaquinol 8 + periplasmic protein disulfide isomerase I (oxidized) ReactionCard |
|
| Transports: |
Not Available |
| Metabolites: |
|
| GO Classification: |
| Component |
|---|
| cell part | | membrane | | Function |
|---|
| catalytic activity | | disulfide oxidoreductase activity | | oxidoreductase activity | | oxidoreductase activity, acting on a sulfur group of donors | | protein disulfide oxidoreductase activity |
|
| Gene Properties |
| Locus tag: |
PA0538 |
| Strand: |
- |
| Entrez Gene ID: |
878500 |
| Accession: |
NP_249229.1 |
| GI: |
15595735 |
| Sequence start: |
596970 |
| Sequence End: |
597479 |
| Sequence Length: |
509 |
| Gene Sequence: |
>PA0538
TTGAGCGCTCTCCTCAAGCCCCTCGACAATCGCTTGTTCTGGCCCGCGGTCGCCATCGGCGGCCTGCTGATCCTCGCTTTCGTCCTCTACCTCCAGCACGTGCGCGGCTTCGCGCCCTGCTCGCTGTGCATCTTCATTCGCCTGGACGTGCTCGGCCTGGTGCTCGCCGGGATCGTCGGCAGCCTCGCCCCGCGTTCGCGGATCGCCGGCGGCATCGCCGCGCTGGGCATGCTCGCCGCCAGCCTGGGCGGGATCTACCACGCCTGGTCGCTGGTCGCGGAAGAGAAACTGGCTGCCCAGGGCATGGGCAGTTGCAAGATGTTCATGGGCTTCCCCGAATGGATACCGCTGGATACCTGGCTGCCGCAGGTCTTCCAGCCCGAGGGGCTGTGCGGCGAAGTGGTCTGGACCCTGCTCGGCCAGTCCATGGCGGTCTGGTCGCTGGCGCTGTTCGTGTTCTGCCTGCTGGTCCTGGCGGCCAAGCTGGCGTTCGGCCGCCGCACCGCCTGA |
| Protein Properties |
| Protein Residues: |
169 |
| Protein Molecular Weight: |
18.1 kDa |
| Protein Theoretical pI: |
8.5 |
| Hydropathicity (GRAVY score): |
0.985 |
| Charge at pH 7 (predicted): |
3.31 |
| Protein Sequence: |
>PA0538
MSALLKPLDNRLFWPAVAIGGLLILAFVLYLQHVRGFAPCSLCIFIRLDVLGLVLAGIVGSLAPRSRIAGGIAALGMLAASLGGIYHAWSLVAEEKLAAQGMGSCKMFMGFPEWIPLDTWLPQVFQPEGLCGEVVWTLLGQSMAVWSLALFVFCLLVLAAKLAFGRRTA |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA0538 and its homologs
|



