|
Record Information |
|---|
| Version |
1.0 |
|---|
| Update Date |
1/22/2018 11:54:54 AM |
|---|
|
Metabolite ID | PAMDB120481 |
|---|
|
Identification |
|---|
| Name: |
3-methylbutanol |
|---|
| Description: | An alkyl alcohol that is butan-1-ol substituted by a methyl group at position 3. |
|---|
|
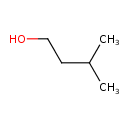
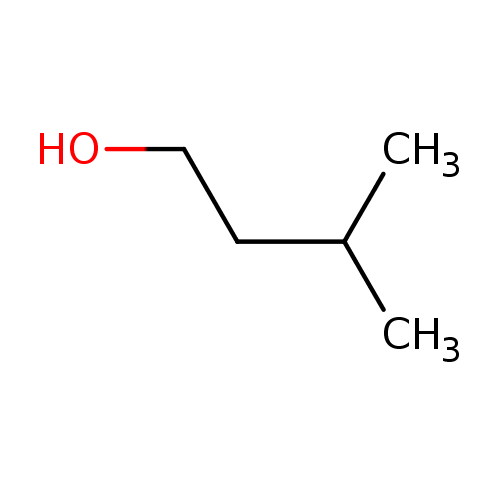
Structure |
|
|---|
| Synonyms: | - 1-HYDROXY-3-METHYLBUTANE
- 2-methyl-4-butanol
- 3-methyl-1-butanol
- 3-Methylbutanol
- 3-methylbutanol
- alcool isoamylique
- i-amyl alcohol
- Iso-amylalkohol
- Isoamyl alcohol
- isoamylol
- isobutylcarbinol
- isopentan-1-ol
- isopentanol
- Isopentyl alcohol
- Isopentylalkohol
- primary isoamyl alcohol
|
|---|
|
Chemical Formula: |
C5H12O |
|---|
| Average Molecular Weight: |
88.149 |
|---|
| Monoisotopic Molecular
Weight: |
88.08881 |
|---|
| InChI Key: |
PHTQWCKDNZKARW-UHFFFAOYSA-N |
|---|
| InChI: | InChI=1S/C5H12O/c1-5(2)3-4-6/h5-6H,3-4H2,1-2H3 |
|---|
| CAS
number: |
123-51-3 |
|---|
| IUPAC Name: | 3-methylbutan-1-ol |
|---|
|
Traditional IUPAC Name: |
isoamyl alcohol |
|---|
| SMILES: | CC(CCO)C |
|---|
|
Chemical Taxonomy |
|---|
|
Taxonomy Description | This compound belongs to the class of organic compounds known as primary alcohols. These are compounds comprising the primary alcohol functional group, with the general structure RCOH (R=alkyl, aryl). |
|---|
|
Kingdom |
Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
|
Class |
Organooxygen compounds |
|---|
| Sub Class | Alcohols and polyols |
|---|
|
Direct Parent |
Primary alcohols |
|---|
| Alternative Parents |
|
|---|
| Substituents |
- Hydrocarbon derivative
- Primary alcohol
- Aliphatic acyclic compound
|
|---|
| Molecular Framework |
Aliphatic acyclic compounds |
|---|
| External Descriptors |
|
|---|
|
Physical Properties |
|---|
| State: |
Liquid |
|---|
| Charge: | 0 |
|---|
|
Melting point: |
-117.2 °C |
|---|
| Experimental Properties: |
| Property | Value | Reference |
|---|
| Melting Point | -117.2 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 26.7 mg/mL at 25 °C | Not Available | | LogP | 1.16 | HANSCH,C ET AL. (1995) |
|
|---|
| Predicted Properties |
|
|---|
|
Biological Properties |
|---|
| Cellular Locations: |
Not Available |
|---|
| Reactions: | |
|---|
|
Pathways: |
Not Available |
|---|
|
Spectra |
|---|
| Spectra: |
| Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS | Not Available |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00dr-9000000000-7bbd6dbb0b9915076ca1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00di-9000000000-c74cb455e67c059f399c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0ab9-9000000000-f827e13ae4abb1b14fbb | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00dr-9000000000-7bbd6dbb0b9915076ca1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00di-9000000000-c74cb455e67c059f399c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0ab9-9000000000-f827e13ae4abb1b14fbb | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-9000000000-7264596be9d872886688 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4r-9000000000-cc98e6ceeea595509f4c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0aor-9000000000-c1813a13047771c3400a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-9000000000-7264596be9d872886688 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4r-9000000000-cc98e6ceeea595509f4c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0aor-9000000000-c1813a13047771c3400a | View in MoNA |
|---|
| MS | Mass Spectrum (Electron Ionization) | splash10-052f-9000000000-370bf1c231476fa54243 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available |
|---|
|
|---|
|
References |
|---|
| References: |
- Zhu FM, Du B, Li J (2014)Aroma enhancement and enzymolysis regulation of grape wine using ß-glycosidase. Food science & nutrition 2, Pubmed: 24804072
- Dourtoglou V, Antonopoulos A, Dourtoglou T, Lalas S (2014)Discrimination of varietal wines according to their volatiles. Food chemistry 159, Pubmed: 24767042
- Dong L, Hou Y, Li F, Piao Y, Zhang X, Zhang X, Li C, Zhao C (2015)Characterization of volatile aroma compounds in different brewing barley cultivars. Journal of the science of food and agriculture 95, Pubmed: 24862930
- Park HJ, Lee SM, Song SH, Kim YS (2013)Characterization of volatile components in makgeolli, a traditional Korean rice wine, with or without pasteurization, during storage. Molecules (Basel, Switzerland) 18, Pubmed: 23698045
- Filannino P, Cardinali G, Rizzello CG, Buchin S, De Angelis M, Gobbetti M, Di Cagno R (2014)Metabolic responses of Lactobacillus plantarum strains during fermentation and storage of vegetable and fruit juices. Applied and environmental microbiology 80, Pubmed: 24487533
|
|---|
| Synthesis Reference: |
Not Available |
|---|
| Material Safety Data Sheet (MSDS) |
Not Available |
|---|
|
Links |
|---|
| External Links: |
|
|---|