|
Record Information |
|---|
| Version |
1.0 |
|---|
| Update Date |
1/22/2018 11:54:54 AM |
|---|
|
Metabolite ID | PAMDB004153 |
|---|
|
Identification |
|---|
| Name: |
Glycolate |
|---|
| Description: | A 2-hydroxy monocarboxylic acid that is acetic acid where the methyl group has been hydroxylated |
|---|
|
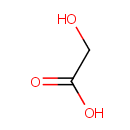
Structure |
|
|---|
| Synonyms: | - 2-Hydroxyacetate
- 2-Hydroxyacetic acid
- a-Hydroxyacetate
- a-Hydroxyacetic acid
- alpha-Hydroxyacetate
- alpha-Hydroxyacetic acid
- Glycocide
- Glycolate
- Glycolic acid
- Glycollate
- Glycollic acid
- GlyPure
- GlyPure 70
- Hydroxyacetate
- Hydroxyacetic acid
- Hydroxyethanoate
- Hydroxyethanoic acid
- Sodium glycolate
- Sodium glycolic acid
- α-Hydroxyacetate
- α-Hydroxyacetic acid
|
|---|
|
Chemical Formula: |
C2H4O3 |
|---|
| Average Molecular Weight: |
76.0514 |
|---|
| Monoisotopic Molecular
Weight: |
76.016043994 |
|---|
| InChI Key: |
AEMRFAOFKBGASW-UHFFFAOYSA-N |
|---|
| InChI: | InChI=1S/C2H4O3/c3-1-2(4)5/h3H,1H2,(H,4,5) |
|---|
| CAS
number: |
79-14-1 |
|---|
| IUPAC Name: | 2-hydroxyacetic acid |
|---|
|
Traditional IUPAC Name: |
glycolic acid |
|---|
| SMILES: | OCC(O)=O |
|---|
|
Chemical Taxonomy |
|---|
|
Taxonomy Description | This compound belongs to the class of organic compounds known as alpha hydroxy acids and derivatives. These are organic compounds containing a carboxylic acid substituted with a hydroxyl group on the adjacent carbon. |
|---|
|
Kingdom |
Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
|
Class |
Hydroxy acids and derivatives |
|---|
| Sub Class | Alpha hydroxy acids and derivatives |
|---|
|
Direct Parent |
Alpha hydroxy acids and derivatives |
|---|
| Alternative Parents |
|
|---|
| Substituents |
- Alpha-hydroxy acid
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Hydrocarbon derivative
- Primary alcohol
- Organooxygen compound
- Carbonyl group
- Alcohol
- Aliphatic acyclic compound
|
|---|
| Molecular Framework |
Aliphatic acyclic compounds |
|---|
| External Descriptors |
|
|---|
|
Physical Properties |
|---|
| State: |
Not Available |
|---|
| Charge: | -1 |
|---|
|
Melting point: |
Not Available |
|---|
| Experimental Properties: |
|
|---|
| Predicted Properties |
|
|---|
|
Biological Properties |
|---|
| Cellular Locations: |
Cytoplasm |
|---|
| Reactions: | |
|---|
|
Pathways: |
- Glycine, serine and threonine metabolism pae00260
- Glyoxylate and dicarboxylate metabolism pae00630
- Metabolic pathways pae01100
- Microbial metabolism in diverse environments pae01120
- Pentose phosphate pathway pae00030
- Pyruvate metabolism pae00620
|
|---|
|
Spectra |
|---|
| Spectra: |
| Spectrum Type | Description | Splash Key | |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) | splash10-0002-0900000000-ed8b8e4a9e2556ea02e2 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (2 TMS) | splash10-00dj-9600000000-8bafc88c7bf4e90fb5e8 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-MS (2 TMS) | splash10-003r-2910000000-bd50bf5bab6f5327eaf4 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-MS | Not Available |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Negative (Annotated) | splash10-004i-9000000000-e942bdae1d60e5f5d649 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Negative (Annotated) | splash10-00di-9000000000-f225de2de3540c3f50a4 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Negative (Annotated) | splash10-00di-9000000000-7de217d97b44f53aad82 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative | splash10-00di-9000000000-88af2b259f82cd1d8938 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative | splash10-004i-9000000000-c968a24f0640b154325b | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative | splash10-0059-9000000000-1dfacf30bf94ce3bf8bb | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004i-9000000000-d961c3c14ec415e3141e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a6r-9000000000-67f73be970ba9f885c4a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-9000000000-f2ccf0b88e0ad65ed4c6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-9000000000-7445713a5fe347bbc8b8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9000000000-26e13242443efc1aa846 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-9000000000-6ba976b949118cd0a86a | View in MoNA |
|---|
| MS | Mass Spectrum (Electron Ionization) | splash10-001i-9000000000-2885890e3bb8c015742f | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available |
|---|
| 2D NMR | [1H,13C] 2D NMR Spectrum | Not Available |
|---|
|
|---|
|
References |
|---|
| References: |
Not Available |
|---|
| Synthesis Reference: |
Not Available |
|---|
| Material Safety Data Sheet (MSDS) |
Not Available |
|---|
|
Links |
|---|
| External Links: |
|
|---|