|
Record Information |
|---|
| Version |
1.0 |
|---|
| Update Date |
1/22/2018 11:54:54 AM |
|---|
|
Metabolite ID | PAMDB001023 |
|---|
|
Identification |
|---|
| Name: |
N-Acetyl-D-muramoate |
|---|
| Description: | N-acetyl-D-muramoate is a member of the chemical class known as Sugar Acids and Derivatives. These are compounds containing a saccharide unit which bears a carboxylic acid group. UDP-N-acetylmuramic acid (UDP-MurNAc) is a precursor for peptidoglycan biosynthesis in bacteria. (PMID 14659546) Peptidoglycan is the major constituent of the cell wall, which is comprised of backbone repeats of N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM). (PMID 18700763) UDP-N-acetylmuramic acid (UDP-MurNAc) is a potent inhibitor of MurA (enolpyruvyl-UDP-GlcNAc synthase). (PMID 15751977) The bacterial cell wall is a polymer consisting of alternating N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) units, cross-linked via peptides appended to MurNAc. (PMID 12369847). The peptidoglycan synthesis pathway starts at the cytoplasm, where in six steps the peptidoglycan precursor a UDP-N-acetylmuramoyl-pentapeptide is synthesized. This precursor is then attached to the memberane acceptor all-trans-undecaprenyl phosphate, generating a N-acetylmuramoyl-pentapeptide-diphosphoundecaprenol, also known as lipid I. Another transferase then adds UDP-N-acetyl-alpha-D-glucosamine, yielding the complete monomeric unit a lipid , also known as lipid . This final lipid intermediate is transferred through the membrane. The peptidoglycan monomers are then polymerized on the outside surface by glycosyltransferases, which form the linear glycan chains, and transpeptidases, which catalyze the formation of peptide crosslinks. |
|---|
|
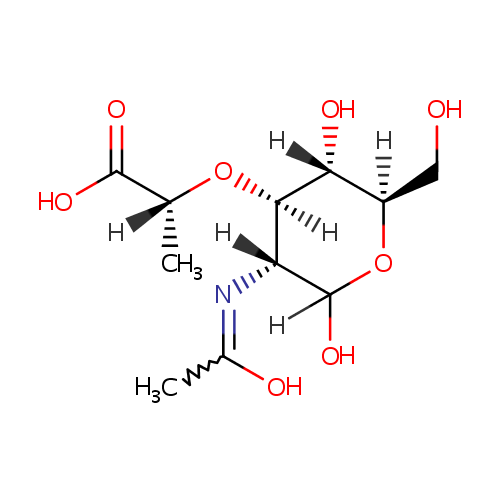
Structure |
|
|---|
| Synonyms: | - N-Acetyl-D-muramoic acid
- N-Acetylmuramate
- N-Acetylmuramic acid
|
|---|
|
Chemical Formula: |
C11H19NO8 |
|---|
| Average Molecular Weight: |
293.2705 |
|---|
| Monoisotopic Molecular
Weight: |
293.111066589 |
|---|
| InChI Key: |
MNLRQHMNZILYPY-MKFCKLDKSA-N |
|---|
| InChI: | InChI=1S/C11H19NO8/c1-4(10(16)17)19-9-7(12-5(2)14)11(18)20-6(3-13)8(9)15/h4,6-9,11,13,15,18H,3H2,1-2H3,(H,12,14)(H,16,17)/t4-,6-,7-,8-,9-,11?/m1/s1 |
|---|
| CAS
number: |
61633-75-8 |
|---|
| IUPAC Name: | (2R)-2-{[(3R,4R,5S,6R)-2,5-dihydroxy-3-[(1-hydroxyethylidene)amino]-6-(hydroxymethyl)oxan-4-yl]oxy}propanoic acid |
|---|
|
Traditional IUPAC Name: |
(2R)-2-{[(3R,4R,5S,6R)-2,5-dihydroxy-3-[(1-hydroxyethylidene)amino]-6-(hydroxymethyl)oxan-4-yl]oxy}propanoic acid |
|---|
| SMILES: | [H][C@](C)(O[C@@]1([H])[C@]([H])(O)[C@@]([H])(CO)OC([H])(O)[C@]1([H])N=C(C)O)C(O)=O |
|---|
|
Chemical Taxonomy |
|---|
|
Taxonomy Description | This compound belongs to the class of organic compounds known as n-acyl-alpha-hexosamines. These are carbohydrate derivatives containing a hexose moiety in which the oxygen atom is replaced by an n-acyl group. |
|---|
|
Kingdom |
Organic compounds |
|---|
| Super Class | Organooxygen compounds |
|---|
|
Class |
Carbohydrates and carbohydrate conjugates |
|---|
| Sub Class | Aminosaccharides |
|---|
|
Direct Parent |
N-acyl-alpha-hexosamines |
|---|
| Alternative Parents |
|
|---|
| Substituents |
- Muramic_acid
- N-acyl-alpha-hexosamine
- Glucosamine
- Amino sugar
- Sugar acid
- Oxane
- Monosaccharide
- Secondary alcohol
- Hemiacetal
- Oxacycle
- Organoheterocyclic compound
- Organic 1,3-dipolar compound
- Propargyl-type 1,3-dipolar organic compound
- Monocarboxylic acid or derivatives
- Ether
- Dialkyl ether
- Carboxylic acid
- Carboxylic acid derivative
- Carboximidic acid derivative
- Carboximidic acid
- Hydrocarbon derivative
- Primary alcohol
- Organonitrogen compound
- Carbonyl group
- Alcohol
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework |
Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors |
|
|---|
|
Physical Properties |
|---|
| State: |
Not Available |
|---|
| Charge: | -1 |
|---|
|
Melting point: |
Not Available |
|---|
| Experimental Properties: |
|
|---|
| Predicted Properties |
|
|---|
|
Biological Properties |
|---|
| Cellular Locations: |
Cytoplasm |
|---|
| Reactions: | |
|---|
|
Pathways: |
- Amino sugar and nucleotide sugar metabolism pae00520
- Phosphotransferase system (PTS) pae02060
|
|---|
|
Spectra |
|---|
| Spectra: |
|
|---|
|
References |
|---|
| References: |
- Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
- Mizyed, S., Oddone, A., Byczynski, B., Hughes, D. W., Berti, P. J. (2005). "UDP-N-acetylmuramic acid (UDP-MurNAc) is a potent inhibitor of MurA (enolpyruvyl-UDP-GlcNAc synthase)." Biochemistry 44:4011-4017. Pubmed: 15751977
- Raymond, J. B., Price, N. P., Pavelka, M. S. Jr (2003). "A method for the enzymatic synthesis and HPLC purification of the peptidoglycan precursor UDP-N-acetylmuramic acid." FEMS Microbiol Lett 229:83-89. Pubmed: 14659546
- Schwartz, B., Markwalder, J. A., Seitz, S. P., Wang, Y., Stein, R. L. (2002). "A kinetic characterization of the glycosyltransferase activity of Eschericia coli PBP1b and development of a continuous fluorescence assay." Biochemistry 41:12552-12561. Pubmed: 12369847
- Suvorov, M., Lee, M., Hesek, D., Boggess, B., Mobashery, S. (2008). "Lytic transglycosylase MltB of Escherichia coli and its role in recycling of peptidoglycan strands of bacterial cell wall." J Am Chem Soc 130:11878-11879. Pubmed: 18700763
- van der Werf, M. J., Overkamp, K. M., Muilwijk, B., Coulier, L., Hankemeier, T. (2007). "Microbial metabolomics: toward a platform with full metabolome coverage." Anal Biochem 370:17-25. Pubmed: 17765195
|
|---|
| Synthesis Reference: |
Not Available |
|---|
| Material Safety Data Sheet (MSDS) |
Not Available |
|---|
|
Links |
|---|
| External Links: |
|
|---|