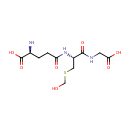
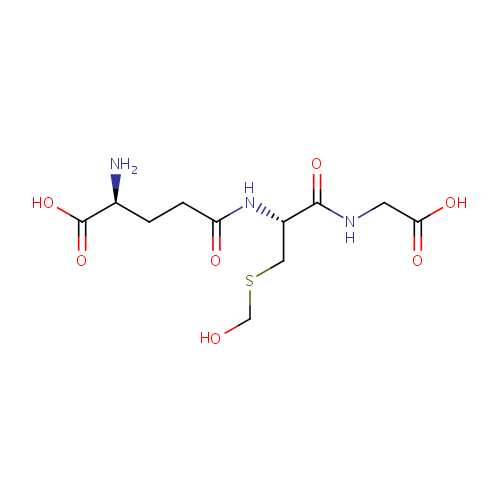
| InChI: | InChI=1S/C11H19N3O7S/c12-6(11(20)21)1-2-8(16)14-7(4-22-5-15)10(19)13-3-9(17)18/h6-7,15H,1-5,12H2,(H,13,19)(H,14,16)(H,17,18)(H,20,21)/t6-,7-/m0/s1 |
|---|
| References: |
- Danielsson O, Shafqat J, Estonius M, el-Ahmad M, Jornvall H: Isozyme multiplicity with anomalous dimer patterns in a class III alcohol dehydrogenase. Effects on the activity and quaternary structure of residue exchanges at "nonfunctional" sites in a native protein. Biochemistry. 1996 Nov 19;35(46):14561-8. Pubmed: 8931553
- Gutheil, W. G., Holmquist, B., Vallee, B. L. (1992). "Purification, characterization, and partial sequence of the glutathione-dependent formaldehyde dehydrogenase from Escherichia coli: a class III alcohol dehydrogenase." Biochemistry 31:475-481. Pubmed: 1731906
- Holmquist B, Moulis JM, Engeland K, Vallee BL: Role of arginine 115 in fatty acid activation and formaldehyde dehydrogenase activity of human class III alcohol dehydrogenase. Biochemistry. 1993 May 18;32(19):5139-44. Pubmed: 8494891
- Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
- Keseler, I. M., Collado-Vides, J., Santos-Zavaleta, A., Peralta-Gil, M., Gama-Castro, S., Muniz-Rascado, L., Bonavides-Martinez, C., Paley, S., Krummenacker, M., Altman, T., Kaipa, P., Spaulding, A., Pacheco, J., Latendresse, M., Fulcher, C., Sarker, M., Shearer, A. G., Mackie, A., Paulsen, I., Gunsalus, R. P., Karp, P. D. (2011). "EcoCyc: a comprehensive database of Escherichia coli biology." Nucleic Acids Res 39:D583-D590. Pubmed: 21097882
- Koivusalo M, Baumann M, Uotila L: Evidence for the identity of glutathione-dependent formaldehyde dehydrogenase and class III alcohol dehydrogenase. FEBS Lett. 1989 Oct 23;257(1):105-9. Pubmed: 2806555
- Lee SL, Wang MF, Lee AI, Yin SJ: The metabolic role of human ADH3 functioning as ethanol dehydrogenase. FEBS Lett. 2003 Jun 5;544(1-3):143-7. Pubmed: 12782305
- Sanghani PC, Bosron WF, Hurley TD: Human glutathione-dependent formaldehyde dehydrogenase. Structural changes associated with ternary complex formation. Biochemistry. 2002 Dec 24;41(51):15189-94. Pubmed: 12484756
- Sanghani PC, Stone CL, Ray BD, Pindel EV, Hurley TD, Bosron WF: Kinetic mechanism of human glutathione-dependent formaldehyde dehydrogenase. Biochemistry. 2000 Sep 5;39(35):10720-9. Pubmed: 10978156
- Yang ZN, Bosron WF, Hurley TD: Structure of human chi chi alcohol dehydrogenase: a glutathione-dependent formaldehyde dehydrogenase. J Mol Biol. 1997 Jan 24;265(3):330-43. Pubmed: 9018047
|
|---|