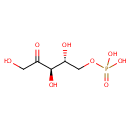
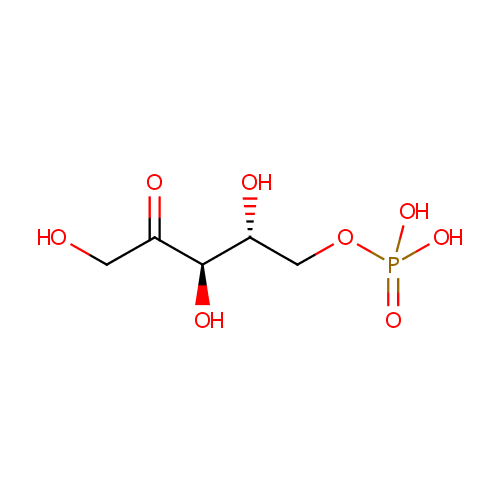
D-Ribulose 5-phosphate (PAMDB000638)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000638 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | D-Ribulose 5-phosphate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | D-Ribulose 5-phosphate is a metabolite in the Pentose phosphate pathway. It is also involved in pentose and glucuronate interconversions, and in Riboflavin metabolism (KEGG) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C5H11O8P | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 230.1098 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 230.01915384 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | FNZLKVNUWIIPSJ-UHNVWZDZSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C5H11O8P/c6-1-3(7)5(9)4(8)2-13-14(10,11)12/h4-6,8-9H,1-2H2,(H2,10,11,12)/t4-,5+/m1/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 4151-19-3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | {[(2R,3R)-2,3,5-trihydroxy-4-oxopentyl]oxy}phosphonic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | Ara | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | OCC(=O)[C@H](O)[C@H](O)COP(O)(O)=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as monosaccharide phosphates. These are monosaccharides comprising a phosphated group linked to the carbohydrate unit. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organooxygen compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Carbohydrates and carbohydrate conjugates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Monosaccharides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Monosaccharide phosphates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | D-Ribose-5-phosphate <> D-Ribulose 5-phosphate D-Ribulose 5-phosphate <> D-Arabinose 5-phosphate D-Ribulose 5-phosphate <> Xylulose 5-phosphate 6-Phosphogluconic acid + NADP <> Carbon dioxide + NADPH + D-Ribulose 5-phosphate + Hydrogen ion D-Ribulose 5-phosphate <> 3,4-Dihydroxy-2-butanone-4-P + Formic acid + Hydrogen ion Adenosine triphosphate + D-Ribulose 5-phosphate + D-Ribulose 5-phosphate <> ADP + D-Ribulose 1,5-bisphosphate NAD(P)<sup>+</sup> + 6-Phosphogluconic acid > NAD(P)H + D-Ribulose 5-phosphate + Carbon dioxide 6-Phosphogluconic acid + NADP > D-Ribulose 5-phosphate + Carbon dioxide + NADPH Adenosine triphosphate + D-Ribulose 5-phosphate > ADP + D-Ribulose 1,5-bisphosphate D-Ribulose 5-phosphate > Formic acid + 1-Deoxy-L-glycero-tetrulose 4-phosphate Adenosine triphosphate + L-Ribulose + D-Ribulose <> ADP + L-Ribulose 5-phosphate + D-Ribulose 5-phosphate 6-Phosphogluconic acid + NADP > D-Ribulose 5-phosphate + Carbon dioxide + NADPH + NADPH D-Ribulose 5-phosphate <> Xylulose 5-phosphate + Xylulose 5-phosphate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Wong, Chi-Huey; McCurry, Stephen D.; Whitesides, George M. Practical enzymic syntheses of ribulose 1,5 bisphosphate and ribose 5-phosphate. Journal of the American Chemical Society (1980), 102(27), 7938-9. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Download (PDF) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in 3,4-dihydroxy-2-butanone-4-phosphate synthase activity
- Specific function:
- Catalyzes the conversion of D-ribulose 5-phosphate to formate and 3,4-dihydroxy-2-butanone 4-phosphate
- Gene Name:
- ribB
- Locus Tag:
- PA4054
- Molecular weight:
- 39.4 kDa
Reactions
| D-ribulose 5-phosphate = formate + L-3,4-dihydroxybutan-2-one 4-phosphate. |
- General function:
- Involved in ribose-5-phosphate isomerase activity
- Specific function:
- D-ribose 5-phosphate = D-ribulose 5-phosphate
- Gene Name:
- rpiA
- Locus Tag:
- PA0330
- Molecular weight:
- 23.7 kDa
Reactions
| D-ribose 5-phosphate = D-ribulose 5-phosphate. |
- General function:
- Involved in catalytic activity
- Specific function:
- D-ribulose 5-phosphate = D-xylulose 5- phosphate
- Gene Name:
- rpe
- Locus Tag:
- PA0607
- Molecular weight:
- 24.1 kDa
Reactions
| D-ribulose 5-phosphate = D-xylulose 5-phosphate. |
- General function:
- Involved in protein binding
- Specific function:
- Catalyzes the interconversion of D-arabinose 5-phosphate and D-ribulose 5-phosphate
- Gene Name:
- kdsD
- Locus Tag:
- PA4457
- Molecular weight:
- 34.2 kDa
Reactions
| D-arabinose 5-phosphate = D-ribulose 5-phosphate. |