|
Record Information |
|---|
| Version |
1.0 |
|---|
| Update Date |
1/22/2018 11:54:54 AM |
|---|
|
Metabolite ID | PAMDB000588 |
|---|
|
Identification |
|---|
| Name: |
2,3-Dihydrodipicolinic acid |
|---|
| Description: | L-2,3-Dihydrodipicolinate is involved in the lysine biosynthesis pathway. L-2,3-Dihydrodipicolinate is produced from a reaction between pyruvate and L-aspartate-semialdehyde, with water as a byproduct. The reaction is catalyzed by dihydrodipicolinate synthase. |
|---|
|
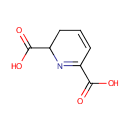
Structure |
|
|---|
| Synonyms: | - 2,3-Di-H-Dipicolinate
- 2,3-Di-H-Dipicolinic acid
- 2,3-Dihydrodipicolinate
- 2,3-DPA
- Dihydrodipicolinate
- Dihydrodipicolinic acid
- L-2,3-Dihydrodipicolinate
- L-2,3-Dihydrodipicolinic acid
|
|---|
|
Chemical Formula: |
C7H7NO4 |
|---|
| Average Molecular Weight: |
169.1348 |
|---|
| Monoisotopic Molecular
Weight: |
169.037507717 |
|---|
| InChI Key: |
UWOCFOFVIBZJGH-UHFFFAOYSA-N |
|---|
| InChI: | InChI=1S/C7H7NO4/c9-6(10)4-2-1-3-5(8-4)7(11)12/h1-2,5H,3H2,(H,9,10)(H,11,12) |
|---|
| CAS
number: |
16052-12-3 |
|---|
| IUPAC Name: | 2,3-dihydropyridine-2,6-dicarboxylic acid |
|---|
|
Traditional IUPAC Name: |
2,3-dihydrodipicolinic acid |
|---|
| SMILES: | OC(=O)C1CC=CC(=N1)C(O)=O |
|---|
|
Chemical Taxonomy |
|---|
|
Taxonomy Description | This compound belongs to the class of organic compounds known as alpha amino acids and derivatives. These are amino acids in which the amino group is attached to the carbon atom immediately adjacent to the carboxylate group (alpha carbon), or a derivative thereof. |
|---|
|
Kingdom |
Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
|
Class |
Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
|
Direct Parent |
Alpha amino acids and derivatives |
|---|
| Alternative Parents |
|
|---|
| Substituents |
- Alpha-amino acid or derivatives
- Dihydropyridinecarboxylic acid derivative
- Dihydropyridine
- Hydropyridine
- Dicarboxylic acid or derivatives
- Ketimine
- Azacycle
- Organoheterocyclic compound
- Organic 1,3-dipolar compound
- Propargyl-type 1,3-dipolar organic compound
- Carboxylic acid
- Hydrocarbon derivative
- Organooxygen compound
- Organonitrogen compound
- Imine
- Carbonyl group
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework |
Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors |
Not Available |
|---|
|
Physical Properties |
|---|
| State: |
Solid |
|---|
| Charge: | -2 |
|---|
|
Melting point: |
135.2 |
|---|
| Experimental Properties: |
| Property | Value | Source |
|---|
| Water Solubility: | 0.03 | PhysProp |
|
|---|
| Predicted Properties |
|
|---|
|
Biological Properties |
|---|
| Cellular Locations: |
Cytoplasm |
|---|
| Reactions: | |
|---|
|
Pathways: |
- Lysine biosynthesis pae00300
- Microbial metabolism in diverse environments pae01120
|
|---|
|
Spectra |
|---|
| Spectra: |
|
|---|
|
References |
|---|
| References: |
- Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
- Keseler, I. M., Collado-Vides, J., Santos-Zavaleta, A., Peralta-Gil, M., Gama-Castro, S., Muniz-Rascado, L., Bonavides-Martinez, C., Paley, S., Krummenacker, M., Altman, T., Kaipa, P., Spaulding, A., Pacheco, J., Latendresse, M., Fulcher, C., Sarker, M., Shearer, A. G., Mackie, A., Paulsen, I., Gunsalus, R. P., Karp, P. D. (2011). "EcoCyc: a comprehensive database of Escherichia coli biology." Nucleic Acids Res 39:D583-D590. Pubmed: 21097882
- van der Werf, M. J., Overkamp, K. M., Muilwijk, B., Coulier, L., Hankemeier, T. (2007). "Microbial metabolomics: toward a platform with full metabolome coverage." Anal Biochem 370:17-25. Pubmed: 17765195
|
|---|
| Synthesis Reference: |
Not Available |
|---|
| Material Safety Data Sheet (MSDS) |
Not Available |
|---|
|
Links |
|---|
| External Links: |
|
|---|