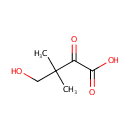
2-Dehydropantoate (PAMDB000576)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000576 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | 2-Dehydropantoate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | 2-dehydropantoate belongs to the class of Branched Fatty Acids. These are fatty acids containing a branched chain. (inferred from compound structure)2-dehydropantoate is invovled in Pantothenate and CoA biosynthesis, and Biosynthesis of secondary metabolites. (KEGG) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C6H9O4 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 145.1333 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 145.050083776 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | PKVVTUWHANFMQC-UHFFFAOYSA-M | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C6H10O4/c1-6(2,3-7)4(8)5(9)10/h7H,3H2,1-2H3,(H,9,10)/p-1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | 4-hydroxy-3,3-dimethyl-2-oxobutanoic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | ketopantoic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | CC(C)(CO)C(=O)C([O-])=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as short-chain keto acids and derivatives. These are keto acids with an alkyl chain the contains less than 6 carbon atoms. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Keto acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Short-chain keto acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Short-chain keto acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | 2-Dehydropantoate + Hydrogen ion + NADPH <> NADP + (R)-Pantoate alpha-Ketoisovaleric acid + Water + 5,10-Methylene-THF + a-Ketoisovaleric acid <> 2-Dehydropantoate + Tetrahydrofolic acid Water + alpha-Ketoisovaleric acid + 5,10-Methylene-THF <> 2-Dehydropantoate + Tetrahydrofolic acid 5,10-Methylene-THF + a-Ketoisovaleric acid + Water > Tetrahydrofolic acid + 2-Dehydropantoate (R)-Pantoate + NADP > 2-Dehydropantoate + NADPH 2-dehydropantoate + NADPH + Hydrogen ion + 2-Dehydropantoate + NADPH > NADP + (R)-pantoate + (R)-Pantoate a-Ketoisovaleric acid + 5,10-Methylene-THF + Water + 5,10-Methylene-THF > Tetrahydrofolic acid + 2-dehydropantoate + Tetrahydrofolic acid + 2-Dehydropantoate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||