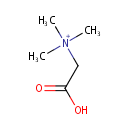
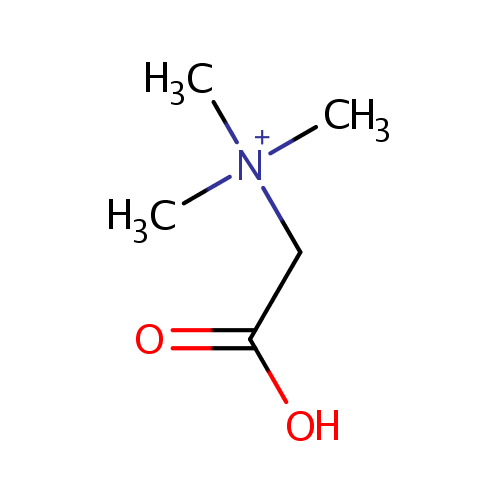
Betaine (PAMDB000564)
| Record Information | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000564 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Betaine | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | Betaine or trimethylglycine is a methylated derivative of glycine. It functions as a methyl donor in that it carries and donates methyl functional groups to facilitate necessary chemical processes. Betaine is an osmoprotectant compound. The accumulation of betaine increases the volume of cytoplasmic water of Pseudomonas aeruginosa and increases the growth rate of osmotically stressed Pseudomonas aeruginosa. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C5H12NO2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 118.1543 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 118.086803633 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | KWIUHFFTVRNATP-UHFFFAOYSA-O | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C5H11NO2/c1-6(2,3)4-5(7)8/h4H2,1-3H3/p+1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 107-43-7 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | (carboxymethyl)trimethylazanium | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | trimethyl glycine | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | C[N+](C)(C)CC(O)=O | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as alpha amino acids. These are amino acids in which the amino group is attached to the carbon atom immediately adjacent to the carboxylate group (alpha carbon). | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic acids and derivatives | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Carboxylic acids and derivatives | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Amino acids, peptides, and analogues | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Alpha amino acids | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | 0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | 293 °C ( Soicke, H., Fitoterapia 1988, V59(1), P73-5); 301 oC dec. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Adenosine triphosphate + Water + Betaine > ADP + Betaine + Hydrogen ion + Phosphate Adenosine triphosphate + Water + Betaine > ADP + Betaine + Hydrogen ion + Phosphate Betaine aldehyde + Water + NADP > Betaine +2 Hydrogen ion + NADPH Betaine aldehyde + Water + NAD <> Betaine +2 Hydrogen ion + NADH Betaine aldehyde + NAD + Water > Betaine + NADH | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Mu, Yun; Guo, Xiao-hui. Improved process for preparation of betaine. Huaxue Yu Shengwu Gongcheng (2005), 22(7), 48-49. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Download (PDF) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in oxidoreductase activity
- Specific function:
- Betaine aldehyde + NAD(+) + H(2)O = betaine + NADH
- Gene Name:
- betB
- Locus Tag:
- PA5373
- Molecular weight:
- 53.3 kDa
Reactions
| Betaine aldehyde + NAD(+) + H(2)O = betaine + NADH. |