Pyridoxal (PAMDB000414)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000414 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
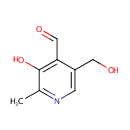
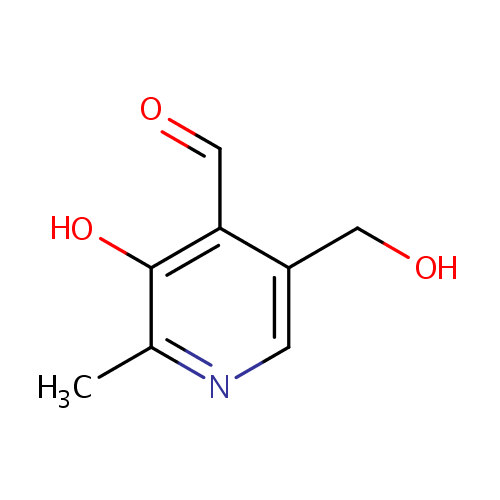
| Name: | Pyridoxal | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | Pyridoxal is the 4-carboxyaldehyde form of vitamin B6 which is converted to pyridoxal phosphate which is a coenzyme for synthesis of amino acids and other compounds. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C8H9NO3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 167.162 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 167.058243159 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | RADKZDMFGJYCBB-UHFFFAOYSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C8H9NO3/c1-5-8(12)7(4-11)6(3-10)2-9-5/h2,4,10,12H,3H2,1H3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 66-72-8 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | 3-hydroxy-5-(hydroxymethyl)-2-methylpyridine-4-carbaldehyde | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | pyridoxal | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | CC1=NC=C(CO)C(C=O)=C1O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as pyridoxals and derivatives. These are compounds containing a pyridoxal moiety, which consists of a pyridine ring substituted at positions 2,3,4, and 5 by a methyl group, a hydroxyl group, a carbaldehyde group, and a hydroxymethyl group, respectively. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organoheterocyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Pyridines and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Pyridine carboxaldehydes | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Pyridoxals and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic heteromonocyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | 0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | 165 °C | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Water + Pyridoxal 5'-phosphate > Phosphate + Pyridoxal Adenosine triphosphate + Pyridoxal <> ADP + Hydrogen ion + Pyridoxal 5'-phosphate Adenosine triphosphate + Pyridoxal <> ADP + Pyridoxal 5'-phosphate Pyridoxamine + Water + Oxygen <> Pyridoxal + Ammonia + Hydrogen peroxide Pyridoxine + Oxygen <> Pyridoxal + Hydrogen peroxide Pyridoxamine + Oxalacetic acid <> Pyridoxal + L-Aspartic acid Pyridoxal 5'-phosphate + Water > Pyridoxal + Inorganic phosphate Pyridoxal + Adenosine triphosphate > Pyridoxal 5'-phosphate + Adenosine diphosphate + ADP | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Fujimoto, Yasuo. Pyridoxal. Jpn. Tokkyo Koho (1971), 2 pp. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Download (PDF) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in pyridoxal kinase activity
- Specific function:
- Phosphorylates B6 vitamers; functions in a salvage pathway. Uses pyridoxamine, but has negligible activity toward pyridoxal and pyridoxine as substrates
- Gene Name:
- pdxY
- Locus Tag:
- PA5516
- Molecular weight:
- 31.3 kDa
Reactions
| ATP + pyridoxal = ADP + pyridoxal 5'-phosphate. |
- General function:
- Coenzyme transport and metabolism
- Specific function:
- Catalyzes the oxidation of either pyridoxine 5'- phosphate (PNP) or pyridoxamine 5'-phosphate (PMP) into pyridoxal 5'-phosphate (PLP)
- Gene Name:
- pdxH
- Locus Tag:
- PA1049
- Molecular weight:
- 24.9 kDa
Reactions
| Pyridoxamine 5'-phosphate + H(2)O + O(2) = pyridoxal 5'-phosphate + NH(3) + H(2)O(2). |
| Pyridoxine 5'-phosphate + O(2) = pyridoxal 5'-phosphate + H(2)O(2). |