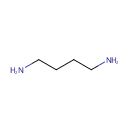
Putrescine (PAMDB000378)
| Record Information | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000378 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Putrescine | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | Putrescine is a polyamine. Putrescine is related to cadaverine (another polyamine). Both are produced by the breakdown of amino acids in living and dead organisms and both are toxic in large doses. Putrescine and cadaverine are largely responsible for the foul odor of putrefying flesh. Putrescine attacks s-adenosyl methionine and converts it to spermidine. Spermidine in turn attacks another s-adenosyl methionine and converts it to spermine. Putrescine is synthesized in small quantities by healthy living cells by the action of ornithine decarboxylase. The polyamines, of which putrescine is one of the simplest, appear to be growth factors necessary for cell division. (Wikipedia) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C4H12N2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 88.1515 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 88.100048394 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | KIDHWZJUCRJVML-UHFFFAOYSA-N | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C4H12N2/c5-3-1-2-4-6/h1-6H2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 110-60-1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | butane-1,4-diamine | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | putrescine | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | NCCCCN | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as monoalkylamines. These are organic compounds containing an primary aliphatic amine group. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organonitrogen compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Amines | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Primary amines | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Monoalkylamines | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | 2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | 27.5 °C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Adenosine triphosphate + Water + Putrescine > ADP + Hydrogen ion + Phosphate + Putrescine Adenosine triphosphate + Water + Putrescine > ADP + Hydrogen ion + Phosphate + Putrescine Hydrogen ion + Ornithine + L-Ornithine <> Carbon dioxide + Putrescine + Ethylenediamine S-Adenosylmethioninamine + Putrescine + Ethylenediamine <> 5'-Methylthioadenosine + Hydrogen ion + Spermidine Adenosine triphosphate + L-Glutamate + Putrescine + Ethylenediamine <> ADP + gamma-Glutamyl-L-putrescine + Hydrogen ion + Phosphate Agmatine + Water <> Putrescine + Urea + Ethylenediamine alpha-Ketoglutarate + Putrescine > 4-Aminobutyraldehyde + L-Glutamate Ornithine <> Putrescine + Carbon dioxide Acetyl-CoA + Putrescine <> Coenzyme A + N-Acetylputrescine Agmatine + Water <> Putrescine + Urea S-Adenosylmethioninamine + Putrescine <> 5'-Methylthioadenosine + Spermidine Adenosine triphosphate + L-Glutamate + Putrescine <> ADP + Phosphate + gamma-Glutamyl-L-putrescine -->-->Hydrogen ion + Ornithine > Carbon dioxide + Putrescine Putrescine + Oxoglutaric acid <> 4-Aminobutyraldehyde + L-Glutamate Putrescine + L-Glutamate + Adenosine triphosphate > Hydrogen ion + gamma-Glutamyl-L-putrescine + ADP + Phosphate Putrescine + S-Adenosylmethioninamine > Hydrogen ion + Spermidine + 5'-Methylthioadenosine Putrescine + Oxoglutaric acid > L-Glutamate + 1-Pyrroline + Water Adenosine triphosphate + L-Glutamate + Putrescine > ADP + Inorganic phosphate + gamma-Glutamyl-L-putrescine Putrescine + Adenosine triphosphate + L-Glutamic acid + L-Glutamate > Phosphate + Adenosine diphosphate + Hydrogen ion + gamma-Glutamyl-L-putrescine + ADP More...Putrescine + Oxoglutaric acid > L-Glutamic acid + 4-Aminobutyraldehyde + L-Glutamate Ornithine + Hydrogen ion + Ornithine > Putrescine + Carbon dioxide Putrescine + S-Adenosylmethioninamine > Spermidine + Hydrogen ion + 5'-S-methyl-5'-thioadenosine Putrescine + Oxoglutaric acid <> L-Glutamic acid + 1-Pyrroline + Water + L-Glutamate | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Dudley, H. W.; Thorpe, W. V. Synthesis of N-methylputrescine and of putrescine. Biochemical Journal (1925), 19 845-9. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in catalytic activity
- Specific function:
- Catalyzes the production of spermidine from putrescine and decarboxylated S-adenosylmethionine (dcSAM), which serves as an aminopropyl donor
- Gene Name:
- speE
- Locus Tag:
- PA1687
- Molecular weight:
- 32.2 kDa
Reactions
| S-adenosylmethioninamine + putrescine = 5'-S-methyl-5'-thioadenosine + spermidine. |
- General function:
- Involved in carboxy-lyase activity
- Specific function:
- L-ornithine = putrescine + CO(2)
- Gene Name:
- speC
- Locus Tag:
- PA4519
- Molecular weight:
- 43.6 kDa
Reactions
| L-ornithine = putrescine + CO(2). |
- General function:
- Involved in nucleotide binding
- Specific function:
- Part of the ABC transporter complex PotABCD involved in spermidine/putrescine import. Responsible for energy coupling to the transport system
- Gene Name:
- potA
- Locus Tag:
- PA3607
- Molecular weight:
- 40 kDa
Reactions
| ATP + H(2)O + polyamine(Out) = ADP + phosphate + polyamine(In). |
- General function:
- Involved in transporter activity
- Specific function:
- Required for the activity of the bacterial periplasmic transport system of putrescine and spermidine
- Gene Name:
- potB
- Locus Tag:
- PA3608
- Molecular weight:
- 32.7 kDa
- General function:
- Involved in transporter activity
- Specific function:
- Required for the activity of the bacterial periplasmic transport system of putrescine and spermidine
- Gene Name:
- potC
- Locus Tag:
- PA3609
- Molecular weight:
- 27.7 kDa
- General function:
- Involved in transporter activity
- Specific function:
- Required for the activity of the bacterial periplasmic transport system of putrescine
- Gene Name:
- potI
- Locus Tag:
- PA0304
- Molecular weight:
- 31.9 kDa
- General function:
- Involved in transporter activity
- Specific function:
- Required for the activity of the bacterial periplasmic transport system of putrescine
- Gene Name:
- potH
- Locus Tag:
- PA0303
- Molecular weight:
- 32.4 kDa
- General function:
- Amino acid transport and metabolism
- Specific function:
- Part of the binding-protein-dependent transport system for putrescine. Probably responsible for energy coupling to the transport system
- Gene Name:
- potG
- Locus Tag:
- PA0302
- Molecular weight:
- 42.8 kDa
- General function:
- Involved in transporter activity
- Specific function:
- Required for the activity of the bacterial periplasmic transport system of putrescine. Polyamine binding protein
- Gene Name:
- potF
- Locus Tag:
- PA1410
- Molecular weight:
- 40.5 kDa
- General function:
- Involved in transporter activity
- Specific function:
- Required for the activity of the bacterial periplasmic transport system of putrescine and spermidine. Polyamine binding protein
- Gene Name:
- potD
- Locus Tag:
- PA3610
- Molecular weight:
- 39.3 kDa
Transporters
- General function:
- Involved in nucleotide binding
- Specific function:
- Part of the ABC transporter complex PotABCD involved in spermidine/putrescine import. Responsible for energy coupling to the transport system
- Gene Name:
- potA
- Locus Tag:
- PA3607
- Molecular weight:
- 40 kDa
Reactions
| ATP + H(2)O + polyamine(Out) = ADP + phosphate + polyamine(In). |
- General function:
- Involved in transporter activity
- Specific function:
- Required for the activity of the bacterial periplasmic transport system of putrescine and spermidine
- Gene Name:
- potB
- Locus Tag:
- PA3608
- Molecular weight:
- 32.7 kDa
- General function:
- Involved in transporter activity
- Specific function:
- Required for the activity of the bacterial periplasmic transport system of putrescine and spermidine
- Gene Name:
- potC
- Locus Tag:
- PA3609
- Molecular weight:
- 27.7 kDa
- General function:
- Involved in transporter activity
- Specific function:
- Required for the activity of the bacterial periplasmic transport system of putrescine
- Gene Name:
- potI
- Locus Tag:
- PA0304
- Molecular weight:
- 31.9 kDa
- General function:
- Involved in transporter activity
- Specific function:
- Required for the activity of the bacterial periplasmic transport system of putrescine
- Gene Name:
- potH
- Locus Tag:
- PA0303
- Molecular weight:
- 32.4 kDa
- General function:
- Involved in transport
- Specific function:
- Imports putrescine
- Gene Name:
- puuP
- Locus Tag:
- PA2041
- Molecular weight:
- 50.1 kDa
- General function:
- Amino acid transport and metabolism
- Specific function:
- Part of the binding-protein-dependent transport system for putrescine. Probably responsible for energy coupling to the transport system
- Gene Name:
- potG
- Locus Tag:
- PA0302
- Molecular weight:
- 42.8 kDa
- General function:
- Involved in transporter activity
- Specific function:
- Required for the activity of the bacterial periplasmic transport system of putrescine. Polyamine binding protein
- Gene Name:
- potF
- Locus Tag:
- PA1410
- Molecular weight:
- 40.5 kDa
- General function:
- Involved in transporter activity
- Specific function:
- Required for the activity of the bacterial periplasmic transport system of putrescine and spermidine. Polyamine binding protein
- Gene Name:
- potD
- Locus Tag:
- PA3610
- Molecular weight:
- 39.3 kDa