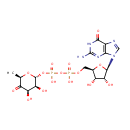
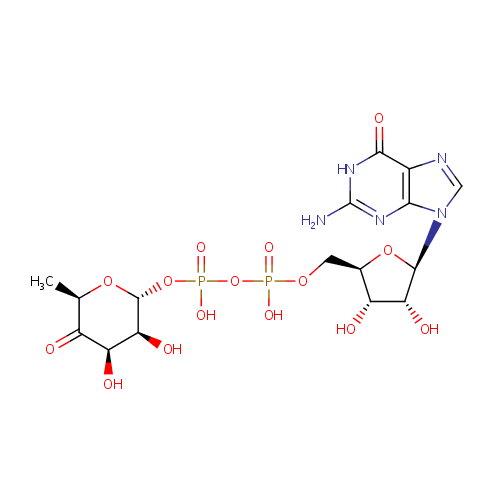
GDP-4-Dehydro-6-deoxy-D-mannose (PAMDB000347)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000347 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | GDP-4-Dehydro-6-deoxy-D-mannose | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | GDP-4-Dehydro-6-deoxy-D-mannose is an intermediate in fructose and mannose metabolism. GDP-4-Dehydro-6-deoxy-D-mannose is generated by GDP-D-mannose-4,6-dehydratase (GMD). This compound is then converted by the FX protein (GDP-4-keto-6-D-deoxymannose epimerase/GDP-4-keto-6-L-galactose reductase) to GDP-L-fucose. . It is also involved in amino sugar and nucleotide sugar metabolism. (KEGG) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C16H23N5O15P2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 587.3258 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 587.066588115 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | PNHLMHWWFOPQLK-BKUUWRAGSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C16H23N5O15P2/c1-4-7(22)9(24)11(26)15(33-4)35-38(30,31)36-37(28,29)32-2-5-8(23)10(25)14(34-5)21-3-18-6-12(21)19-16(17)20-13(6)27/h3-5,8-11,14-15,23-26H,2H2,1H3,(H,28,29)(H,30,31)(H3,17,19,20,27)/t4-,5-,8-,9+,10-,11+,14-,15-/m1/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 18186-48-6 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | [({[(2R,3S,4R,5R)-5-(2-amino-6-oxo-6,9-dihydro-1H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy]({[(2R,3S,4R,6R)-3,4-dihydroxy-6-methyl-5-oxooxan-2-yl]oxy})phosphinic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | gdp-4-keto-6-deoxymannose | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | C[C@H]1O[C@H](OP(O)(=O)OP(O)(=O)OC[C@H]2O[C@H]([C@H](O)[C@@H]2O)N2C=NC3=C2N=C(N)NC3=O)[C@@H](O)[C@@H](O)C1=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as purine nucleotide sugars. These are purine nucleotides bound to a saccharide derivative through the terminal phosphate group. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Nucleosides, nucleotides, and analogues | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Purine nucleotides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Purine nucleotide sugars | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Purine nucleotide sugars | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic heteropolycyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | GDP-4-Dehydro-6-deoxy-D-mannose > GDP-4-Oxo-L-fucose Guanosine diphosphate mannose <> GDP-4-Dehydro-6-deoxy-D-mannose + Water GDP-L-Fucose + NADP <> GDP-4-Dehydro-6-deoxy-D-mannose + NADPH + Hydrogen ion GDP-L-Fucose + NADP > GDP-4-Dehydro-6-deoxy-D-mannose + NADPH GDP-4-Dehydro-6-deoxy-D-mannose + NADPH + Hydrogen ion + NADPH > GDP-L-Fucose + NADP | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Oths, Philip J.; Mayer, Robert M.; Floss, Heinz G. Stereochemistry and mechanism of the GDP-mannose dehydratase reaction. Carbohydrate Research (1990), 198(1), 91-100. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||