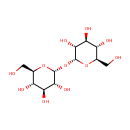
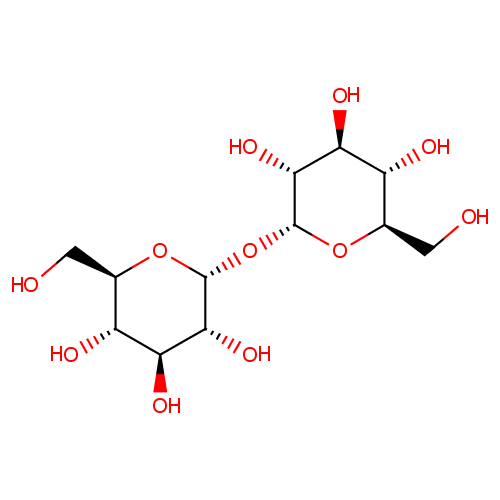
Trehalose (PAMDB000214)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000214 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Trehalose | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | Trehalose, also known as mycose, is a 1-alpha (disaccharide) sugar found extensively but not abundantly in nature. It is thought to be implicated in anhydrobiosis - the ability of plants and animals to withstand prolonged periods of desiccation. The sugar is thought to form a gel phase as cells dehydrate, which prevents disruption of internal cell organelles by effectively splinting them in position. Rehydration then allows normal cellular activity to be resumed without the major, generally lethal damage that would normally follow a dehydration/reyhdration cycle.Trehalose is a non-reducing sugar formed from two glucose units joined by a 1-1 alpha bond giving it the name of alpha-D-glucopyranoglucopyranosyl-1,1-alpha-D-glucopyranoside. The bonding makes trehalose very resistant to acid hydrolysis, and therefore stable in solution at high temperatures even under acidic conditions. The bonding also keeps non-reducing sugars in closed-ring form, such that the aldehyde or ketone end-groups do not bind to the lysine or arginine residues of proteins (a process called glycation). The enzyme trehalase breaks it into two glucose molecules. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C12H22O11 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 342.2965 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 342.116211546 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | HDTRYLNUVZCQOY-LIZSDCNHSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C12H22O11/c13-1-3-5(15)7(17)9(19)11(21-3)23-12-10(20)8(18)6(16)4(2-14)22-12/h3-20H,1-2H2/t3-,4-,5-,6-,7+,8+,9-,10-,11-,12-/m1/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 99-20-7 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | (2R,3S,4S,5R,6R)-2-(hydroxymethyl)-6-{[(2R,3R,4S,5S,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}oxane-3,4,5-triol | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | α,α'-trehalose | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | OC[C@H]1O[C@H](O[C@H]2O[C@H](CO)[C@@H](O)[C@H](O)[C@H]2O)[C@H](O)[C@@H](O)[C@@H]1O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as o-glycosyl compounds. These are glycoside in which a sugar group is bonded through one carbon to another group via a O-glycosidic bond. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organooxygen compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Carbohydrates and carbohydrate conjugates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Glycosyl compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | O-glycosyl compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic heteromonocyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | 0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | 203 °C | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Phosphoenolpyruvic acid + Trehalose > Pyruvic acid + Trehalose 6-phosphate Water + Trehalose 6-phosphate > Phosphate + Trehalose Trehalose + Water <>2 D-Glucose Trehalose + Protein N(pi)-phospho-L-histidine <> Trehalose 6-phosphate + Protein histidine Starch <> Trehalose Trehalose + Water > b-D-Glucose + D-Glucose Trehalose 6-phosphate + Water > Trehalose + Inorganic phosphate Trehalose + Water > b-D-Glucose + alpha-D-Glucose | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Murao, Sawao; Nagano, Hiroto; Ogura, Sei; Nishino, Toyokazu. Enzymic synthesis of trehalose from maltose. Agricultural and Biological Chemistry (1985), 49(7), 2113-18. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Download (PDF) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in catalytic activity
- Specific function:
- Provides the cells with the ability to utilize trehalose at high osmolarity by splitting it into glucose molecules that can subsequently be taken up by the phosphotransferase-mediated uptake system
- Gene Name:
- treA
- Locus Tag:
- PA2416
- Molecular weight:
- 61.2 kDa
Reactions
| Alpha,alpha-trehalose + H(2)O = 2 D-glucose. |