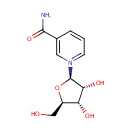
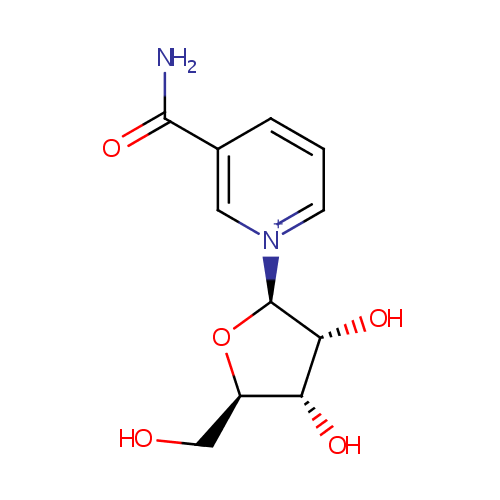
| References: |
- Belenky P, Racette FG, Bogan KL, McClure JM, Smith JS, Brenner C: Nicotinamide riboside promotes Sir2 silencing and extends lifespan via Nrk and Urh1/Pnp1/Meu1 pathways to NAD+. Cell. 2007 May 4;129(3):473-84. Pubmed: 17482543
- Bieganowski, P., Brenner, C. (2004). "Discoveries of nicotinamide riboside as a nutrient and conserved NRK genes establish a Preiss-Handler independent route to NAD+ in fungi and humans." Cell 117:495-502. Pubmed: 15137942
- Bogan KL, Brenner C: Nicotinic acid, nicotinamide, and nicotinamide riboside: a molecular evaluation of NAD+ precursor vitamins in human nutrition. Annu Rev Nutr. 2008;28:115-30. doi: 10.1146/annurev.nutr.28.061807.155443. Pubmed: 18429699
- Canto C, Houtkooper RH, Pirinen E, Youn DY, Oosterveer MH, Cen Y, Fernandez-Marcos PJ, Yamamoto H, Andreux PA, Cettour-Rose P, Gademann K, Rinsch C, Schoonjans K, Sauve AA, Auwerx J: The NAD(+) precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metab. 2012 Jun 6;15(6):838-47. doi: 10.1016/j.cmet.2012.04.022. Pubmed: 22682224
- Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
- Keseler, I. M., Collado-Vides, J., Santos-Zavaleta, A., Peralta-Gil, M., Gama-Castro, S., Muniz-Rascado, L., Bonavides-Martinez, C., Paley, S., Krummenacker, M., Altman, T., Kaipa, P., Spaulding, A., Pacheco, J., Latendresse, M., Fulcher, C., Sarker, M., Shearer, A. G., Mackie, A., Paulsen, I., Gunsalus, R. P., Karp, P. D. (2011). "EcoCyc: a comprehensive database of Escherichia coli biology." Nucleic Acids Res 39:D583-D590. Pubmed: 21097882
- Magni G, Amici A, Emanuelli M, Orsomando G, Raffaelli N, Ruggieri S: Enzymology of NAD+ homeostasis in man. Cell Mol Life Sci. 2004 Jan;61(1):19-34. Pubmed: 14704851
- Pankiewicz KW, Watanabe KA, Lesiak-Watanabe K, Goldstein BM, Jayaram HN: The chemistry of nicotinamide adenine dinucleotide (NAD) analogues containing C-nucleosides related to nicotinamide riboside. Curr Med Chem. 2002 Apr;9(7):733-41. Pubmed: 11966436
- Schalk-Hihi C, Zhang YZ, Markham GD: The conformation of NADH bound to inosine 5'-monophosphate dehydrogenase determined by transferred nuclear Overhauser effect spectroscopy. Biochemistry. 1998 May 19;37(20):7608-16. Pubmed: 9585576
- Wall KA, Klis M, Kornet J, Coyle D, Ame JC, Jacobson MK, Slama JT: Inhibition of the intrinsic NAD+ glycohydrolase activity of CD38 by carbocyclic NAD analogues. Biochem J. 1998 Nov 1;335 ( Pt 3):631-6. Pubmed: 9794804
|
|---|