| Identification |
| Name: |
Phosphoglucosamine mutase |
| Synonyms: |
Not Available |
| Gene Name: |
glmM |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in intramolecular transferase activity, phosphotransferases |
| Specific Function: |
Catalyzes the conversion of glucosamine-6-phosphate to glucosamine-1-phosphate. Can also catalyze the formation of glucose-6-P from glucose-1-P, although at a 1400-fold lower rate |
| Cellular Location: |
Not Available |
| KEGG Pathways: |
- Amino sugar and nucleotide sugar metabolism pae00520
- Lipopolysaccharide biosynthesis pae00540
- Metabolic pathways pae01100
|
| KEGG Reactions: |
|
alpha-D-Glucosamine 1-phosphate | + | 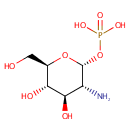 | ↔ | 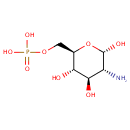 |
| |
|
| SMPDB Reactions: |
|
 | ↔ | Glucosamine-1P | + | 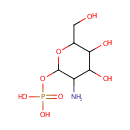 |
| | |
D-glucosamine 6-phosphate | ↔ |  |
| |
|
| PseudoCyc/BioCyc Reactions: |
|
 | ↔ |  |
| |
|
| Complex Reactions: |
|
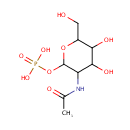 | ↔ |  |
| |
|
| Transports: |
Not Available |
| Metabolites: |
| PAMDB ID | Name | View |
|---|
| PAMDB002052 | alpha-D-Glucosamine 1-phosphate | MetaboCard | | PAMDB000508 | alpha-D-Glucose 1,6-bisphosphate | MetaboCard | | PAMDB002053 | beta-D-Fructose 2-phosphate | MetaboCard | | PAMDB000310 | Glucosamine 6-phosphate | MetaboCard | | PAMDB000256 | Glucosamine-1P | MetaboCard | | PAMDB000357 | N-Acetyl-glucosamine 1-phosphate | MetaboCard |
|
| GO Classification: |
| Function |
|---|
| binding | | catalytic activity | | cation binding | | intramolecular transferase activity | | intramolecular transferase activity, phosphotransferases | | ion binding | | isomerase activity | | magnesium ion binding | | metal ion binding | | phosphoglucosamine mutase activity | | Process |
|---|
| carbohydrate metabolic process | | metabolic process | | primary metabolic process |
|
| Gene Properties |
| Locus tag: |
PA4749 |
| Strand: |
- |
| Entrez Gene ID: |
881713 |
| Accession: |
NP_253437.1 |
| GI: |
15599943 |
| Sequence start: |
5333567 |
| Sequence End: |
5334904 |
| Sequence Length: |
1337 |
| Gene Sequence: |
>PA4749
ATGAGCAGAAAATACTTCGGGACTGACGGTATCCGTGGTCGTGTTGGCGAGTTTCCGATTACTCCGGACTTCGTTCTCAAGCTGGGCTGGGCGGTGGGCATGGCCTTCCGCCGGCAGGGCAACTGCCGGGTCCTGATCGGCAAGGACACGCGCAGCTCGGGTTACATGTTCGAGTCGGCCTTCGAGGCCGGGCTCTCCGCTTCCGGCGCCGATACCCTGTTGCTGGGACCGATGCCTACCCCCGGCATCGCCTACCTGACGCGGACTTTCCACGCCGAGGCCGGGGTGGTGATCAGCGCTTCGCACAATCCCCACGACGACAACGGCATCAAGTTCTTCTCCGGGCAGGGGACCAAGCTGCCGGACGACGTCGAGCTGATGATCGAGGAACTGCTCGATGCGCCGATGACCGTGGTGGAGTCCGCCCGTCTCGGCAAGGTTTCGCGGATCAATGACGCGGCGGGGCGCTACATCGAATTCTGCAAGAGCAGCGTGCCGACCAGCACCGACTTCAACGGTCTCAAGGTCGTGCTCGACTGCGCCAACGGCGCTACCTACAAGATCGCTCCCAGCGTCTTCCGGGAACTGGGCGCGGAAGTGACGGTGCTGGCGGCCAGCCCGAATGGCCTGAACATCAACGACAAGTGCGGTTCCACTCACCTGGATGGCCTGCAGGCTGCGGTGGTCGAGCACCATGCCGACCTCGGCATCGCCTTCGACGGCGACGGCGACCGGGTGATGATGGTCGACCATACCGGTGCGGTCGTGGATGGCGACGAACTGCTTTTCCTCATCGCGCGCGACCTGCAGGAAAGCGGGCGGCTGCAAGGTGGTGTGGTCGGCACCCTGATGAGCAACCTCGGCCTGGAGCTGGCTTTGCAGGAACTGCATATCCCGTTCGTGCGGGCGAAGGTCGGCGATCGCTATGTGATGGCCGAACTGCTGGCTCGGAACTGGATGCTGGGAGGGGAGAATTCGGGGCATATCGTCTGCTGCCAGAACACCACGACGGGCGATGCCATCATCGCGGCGCTACAGGTGCTGATGGCGCTCAAGCATCGCGGCCAGACCCTGGCAGAGGCGCGCCAGGGGATTCGCAAGTGCCCCCAGGTGTTGATCAATGTGCGCTTCAAGGGCGAAAACGATCCGCTCGAGCACCCCTCGGTGAAGGAGGCCAGCGTCCGCGTCACCGAGCAAATGGGCGGTCGCGGTCGTGTCCTGCTGCGCAAATCCGGCACAGAGCCGTTGGTCCGGGTGATGGTCGAGGGCGACGAGGAAGCTAGCGTTCGTGCCCACGCCGAACAGCTGGCAAAAATTGTTTCTGAGGTATGTGCTTGA |
| Protein Properties |
| Protein Residues: |
445 |
| Protein Molecular Weight: |
47.8 kDa |
| Protein Theoretical pI: |
5.95 |
| Hydropathicity (GRAVY score): |
-0.024 |
| Charge at pH 7 (predicted): |
-7.34 |
| Protein Sequence: |
>PA4749
MSRKYFGTDGIRGRVGEFPITPDFVLKLGWAVGMAFRRQGNCRVLIGKDTRSSGYMFESAFEAGLSASGADTLLLGPMPTPGIAYLTRTFHAEAGVVISASHNPHDDNGIKFFSGQGTKLPDDVELMIEELLDAPMTVVESARLGKVSRINDAAGRYIEFCKSSVPTSTDFNGLKVVLDCANGATYKIAPSVFRELGAEVTVLAASPNGLNINDKCGSTHLDGLQAAVVEHHADLGIAFDGDGDRVMMVDHTGAVVDGDELLFLIARDLQESGRLQGGVVGTLMSNLGLELALQELHIPFVRAKVGDRYVMAELLARNWMLGGENSGHIVCCQNTTTGDAIIAALQVLMALKHRGQTLAEARQGIRKCPQVLINVRFKGENDPLEHPSVKEASVRVTEQMGGRGRVLLRKSGTEPLVRVMVEGDEEASVRAHAEQLAKIVSEVCA |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA4749 and its homologs
|




