| Identification |
| Name: |
Membrane-bound lytic murein transglycosylase B |
| Synonyms: |
- 35 kDa soluble lytic transglycosylase
- Murein hydrolase B
- Slt35
|
| Gene Name: |
mltB |
| Enzyme Class: |
Not Available |
| Biological Properties |
| General Function: |
peptidoglycan biosynthetic process |
| Specific Function: |
lytic transglycosylase activity |
| Cellular Location: |
Cell outer membrane; Lipid-anchor; Periplasmic side |
| KEGG Pathways: |
|
| KEGG Reactions: |
Not Available |
| SMPDB Reactions: |
Not Available |
| PseudoCyc/BioCyc Reactions: |
|
a peptidoglycan polymer | ? | a peptidoglycan polymer with 1,6-anhydromuropeptide end | + | a peptidoglycan polymer with GlcNAc end |
| a peptidoglycan polymer ? a peptidoglycan polymer with 1,6-anhydromuropeptide end + a peptidoglycan polymer with GlcNAc end ReactionCard |
|
| Complex Reactions: |
|
two linked disacharide tetrapeptide and tripeptide murein units (uncrosslinked, middle of chain) | → | 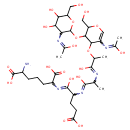 | + | 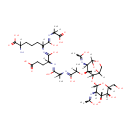 |
| | |
two linked disacharide tripeptide murein units (uncrosslinked, middle of chain) | → | 2  |
| | |
two linked disacharide tetrapeptide murein units (uncrosslinked, middle of chain) | → | 2  |
| | |
three disacharide linked murein units (tetrapeptide crosslinked tetrapeptide (A2pm->D-ala), one uncrosslinked tetrapaptide) (middle of chain) | → |  | + | two disacharide linked murein units, tetrapeptide corsslinked tetrapeptide (A2pm->D-ala) (middle of chain) |
| |
|
| Transports: |
Not Available |
| Metabolites: |
| PAMDB ID | Name | View |
|---|
| PAMDB001657 | N-Acetyl-D-glucosamine(anhydrous)N-Acetylmuramyl-tetrapeptide | MetaboCard | | PAMDB001658 | N-Acetyl-D-glucosamine(anhydrous)N-Acetylmuramyl-tripeptide | MetaboCard |
|
| GO Classification: |
| Function |
|---|
| lytic transglycosylase activity | | Process |
|---|
| peptidoglycan biosynthetic process |
|
| Gene Properties |
| Locus tag: |
PA4444 |
| Strand: |
+ |
| Entrez Gene ID: |
880971 |
| Accession: |
NP_253134.1 |
| GI: |
15599640 |
| Sequence start: |
4977869 |
| Sequence End: |
4978972 |
| Sequence Length: |
1103 |
| Gene Sequence: |
>PA4444
ATGCGCCGTACCGCCCTCGCCCTGCCCCTGTTCCTTCTGGTCTCAGCATGCAGCAGCGAACCGACGCCACCACCGAAACCCGCCGCCAAACCCCAGGCCCGCACCGTCATTTCACCCCGCCCCGTACGCCAGTCGGTGCAACCGATACTGCCGCTGCGCGGCGATTACGCGAACAATCCGGCGGCACAGCACTTCATCGACAGGATGGTCAGCCAGCACGGCTTCAACCGCCAGCAACTGCACGATCTGTTCGCCCAGACCCAGCGCCTGGACTGGGTGATCCGCCTGATGGACCGGCAAGCCCCGACCTATACCCCACCCAGCGGACCGAACGGCGCCTGGCTGCGCTACCGGAAGAAGTTCGTCACGCCAGGCAACGTACAGAACGGCGTGCTGTTCTGGGACCAATACGAAACCGACCTGCAACGGGCATCGCGCGTCTACGGCGTGCCGCCGGAGATCATCGTCGGCATCATCGGCGTGGAAACCCGCTGGGGGCGTGTGATGGGCAAGACGCGGATCATCGATGCGCTGTCCACCCTGTCCTTCTCCTACCCTCGCCGCGCGGAATTCTTCAGCGGCGAACTGGAGCAATTCCTCCTCCAGGCGCGCAAGGAAGGCACCGACCCGCTGGCCCTGCGCGGTTCCTATGCCGGCGCCATGGGCTACGGCCAGTTCATGCCGTCTTCATTCACCAAGTACGCGGTGGACTTCGATGGCGATGGGCATATCGACCTGTGGAATCCGCGTGACGCCATCGGCAGCGTCGCCAACTATTTCAAGCAGCACGGCTGGGTCAGCGGCGATCGCGTGGCGGTTCCCGCCAGTGGCCGGGCTCCCTCGCTGGAAGATGGCTTCAAGACGCTGTACCCGCTGGACGTGCTCGCTTCCGCCGGATTACGCCCGCAGGGTCCGCTCGGCGGCCACCGGCAAGCCAGCCTGCTGCGCCTGGACATGGGCAGGAACTACCAGTACTGGTACGGCCTGCCGAACTTCTACGTGATCACCCGCTATAACCACAGCACCCACTACGCGATGGCCGTCTGGGAACTGGGCAAGGAAGTCGACCGGGTGCGTCACCGCTCCGTCGTCAGGCAGGATTAG |
| Protein Properties |
| Protein Residues: |
367 |
| Protein Molecular Weight: |
41.4 kDa |
| Protein Theoretical pI: |
10.16 |
| Hydropathicity (GRAVY score): |
-0.407 |
| Charge at pH 7 (predicted): |
15.12 |
| Protein Sequence: |
>PA4444
MRRTALALPLFLLVSACSSEPTPPPKPAAKPQARTVISPRPVRQSVQPILPLRGDYANNPAAQHFIDRMVSQHGFNRQQLHDLFAQTQRLDWVIRLMDRQAPTYTPPSGPNGAWLRYRKKFVTPGNVQNGVLFWDQYETDLQRASRVYGVPPEIIVGIIGVETRWGRVMGKTRIIDALSTLSFSYPRRAEFFSGELEQFLLQARKEGTDPLALRGSYAGAMGYGQFMPSSFTKYAVDFDGDGHIDLWNPRDAIGSVANYFKQHGWVSGDRVAVPASGRAPSLEDGFKTLYPLDVLASAGLRPQGPLGGHRQASLLRLDMGRNYQYWYGLPNFYVITRYNHSTHYAMAVWELGKEVDRVRHRSVVRQD |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA4444 and its homologs
|


