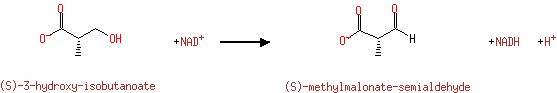| Identification |
| Name: |
3-hydroxyisobutyrate dehydrogenase |
| Synonyms: |
Not Available |
| Gene Name: |
mmsB |
| Enzyme Class: |
Not Available |
| Biological Properties |
| General Function: |
isoleucine catabolic process, leucine catabolic process, carbon utilization, cellular catabolic process, pentose-phosphate shunt, oxidation-reduction process, valine metabolic process |
| Specific Function: |
glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity, phosphogluconate dehydrogenase (decarboxylating) activity, 3-hydroxyisobutyrate dehydrogenase activity, NAD binding, oxidoreductase activity, oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor, coenzyme binding |
| Cellular Location: |
Cytoplasmic |
| KEGG Pathways: |
|
| KEGG Reactions: |
Not Available |
| SMPDB Reactions: |
Not Available |
| PseudoCyc/BioCyc Reactions: |
| ( S)-3-hydroxy-isobutanoate + NAD + → ( S)-methylmalonate-semialdehyde + NADH + H +ReactionCard |
|
| Complex Reactions: |
Not Available |
| Transports: |
Not Available |
| Metabolites: |
Not Available |
| GO Classification: |
| Function |
|---|
| glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity | | phosphogluconate dehydrogenase (decarboxylating) activity | | 3-hydroxyisobutyrate dehydrogenase activity | | NAD binding | | oxidoreductase activity | | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | | coenzyme binding | | Process |
|---|
| isoleucine catabolic process | | leucine catabolic process | | carbon utilization | | cellular catabolic process | | pentose-phosphate shunt | | oxidation-reduction process | | valine metabolic process |
|
| Gene Properties |
| Locus tag: |
PA3569 |
| Strand: |
- |
| Entrez Gene ID: |
879097 |
| Accession: |
NP_252259.1 |
| GI: |
15598765 |
| Sequence start: |
4001196 |
| Sequence End: |
4002092 |
| Sequence Length: |
896 |
| Gene Sequence: |
>PA3569
ATGACCGACATCGCATTCCTCGGCCTGGGCAACATGGGTGGGCCGATGGCCGCCAACCTGCTCAAGGCCGGCCACCGGGTGAATGTCTTCGACTTGCAGCCCAAGGCCGTGCTGGGCCTGGTCGAGCAGGGCGCGCAGGGCGCCGATAGCGCCTTGCAGTGCTGCGAAGGCGCCGAAGTGGTGATCAGCATGCTGCCGGCCGGGCAGCACGTGGAAAGCCTGTATCTCGGCGACGACGGCCTGCTCGCGCGGGTCGCCGGCAAGCCCCTGCTGATCGACTGCTCGACCATCGCCCCGGAGACCGCGCGCAAGGTCGCCGAGGCCGCCGCGGCGAAGGGCCTGACCCTGCTCGACGCGCCGGTTTCCGGCGGCGTCGGCGGCGCCCGCGCCGGCACCCTGAGCTTCATCGTCGGCGGCCCCGCCGAAGGCTTCGCGCGGGCCCGGCCGGTCCTCGAGAACATGGGCCGGAACATCTTCCACGCCGGCGATCACGGCGCCGGCCAGGTGGCGAAGATCTGCAACAACATGCTCCTCGGCATCCTCATGGCCGGCACCGCCGAGGCCCTGGCGCTGGGGGTGAAGAACGGCCTCGACCCGGCGGTGCTGTCCGAGGTGATGAAGCAGAGTTCCGGCGGCAACTGGGCGCTGAACCTCTACAACCCCTGGCCCGGGGTGATGCCGCAGGCGCCGGCGAGCAACGGCTATGCCGGCGGTTTCCAGGTGCGCCTGATGAACAAGGACCTCGGCCTGGCGCTGGCCAACGCCCAGGCGGTGCAGGCCTCGACGCCGCTCGGCGCGCTGGCGCGCAACCTGTTCAGCCTGCACGCCCAGGCCGATGCCGAGCACGAGGGGCTGGACTTCTCCAGCATCCAGAAGCTCTACCGCGGCAAGGACTGA |
| Protein Properties |
| Protein Residues: |
298 |
| Protein Molecular Weight: |
30.5 kDa |
| Protein Theoretical pI: |
6.24 |
| Hydropathicity (GRAVY score): |
0.183 |
| Charge at pH 7 (predicted): |
-2.68 |
| Protein Sequence: |
>PA3569
MTDIAFLGLGNMGGPMAANLLKAGHRVNVFDLQPKAVLGLVEQGAQGADSALQCCEGAEVVISMLPAGQHVESLYLGDDGLLARVAGKPLLIDCSTIAPETARKVAEAAAAKGLTLLDAPVSGGVGGARAGTLSFIVGGPAEGFARARPVLENMGRNIFHAGDHGAGQVAKICNNMLLGILMAGTAEALALGVKNGLDPAVLSEVMKQSSGGNWALNLYNPWPGVMPQAPASNGYAGGFQVRLMNKDLGLALANAQAVQASTPLGALARNLFSLHAQADAEHEGLDFSSIQKLYRGKD |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA3569 and its homologs
|