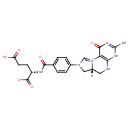
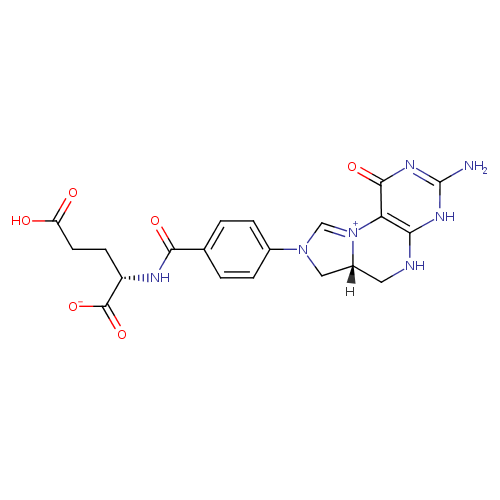
| InChI: | InChI=1S/C20H21N7O6/c21-20-24-16-15(18(31)25-20)27-9-26(8-12(27)7-22-16)11-3-1-10(2-4-11)17(30)23-13(19(32)33)5-6-14(28)29/h1-4,9,12-13H,5-8H2,(H6-,21,22,23,24,25,28,29,30,31,32,33)/p-1/t12-,13+/m1/s1 |
|---|
| References: |
- Field MS, Anderson DD, Stover PJ: Mthfs is an Essential Gene in Mice and a Component of the Purinosome. Front Genet. 2011;2:36. doi: 10.3389/fgene.2011.00036. [22303332 ]
- Oberpichler I, Pierik AJ, Wesslowski J, Pokorny R, Rosen R, Vugman M, Zhang F, Neubauer O, Ron EZ, Batschauer A, Lamparter T: A photolyase-like protein from Agrobacterium tumefaciens with an iron-sulfur cluster. PLoS One. 2011;6(10):e26775. doi: 10.1371/journal.pone.0026775. Epub 2011 Oct 31. [22066008 ]
- Telegina TA, Liudnikova TA, Zemskova IuL, Sviridov EA, Kritskii MS: [Tolerance of 5,10-methenyltetrahydrofolate to ultraviolet radiation]. Prikl Biokhim Mikrobiol. 2005 May-Jun;41(3):315-23. [15977793 ]
- Eadsforth TC, Cameron S, Hunter WN: The crystal structure of Leishmania major N(5),N(10)-methylenetetrahydrofolate dehydrogenase/cyclohydrolase and assessment of a potential drug target. Mol Biochem Parasitol. 2012 Feb;181(2):178-85. doi: 10.1016/j.molbiopara.2011.11.004. Epub 2011 Nov 15. [22108435 ]
- Lin CJ, Wen MJ, Hung YJ, Pei D, Kuo SW, Hsieh CH: The impact of 5,10-methenyltetrahydrofolate synthetase polymorphism on diabetic nephropathy in the Taiwanese population. Genet Test Mol Biomarkers. 2012 Feb;16(2):142-5. doi: 10.1089/gtmb.2011.0050. Epub 2011 Sep 6. [21895484 ]
- Field MS, Szebenyi DM, Stover PJ: Regulation of de novo purine biosynthesis by methenyltetrahydrofolate synthetase in neuroblastoma. J Biol Chem. 2006 Feb 17;281(7):4215-21. Epub 2005 Dec 19. [16365037 ]
- Kruszyna L, Lianeri M, Rydzanicz M, Gajecka M, Szyfter K, Jagodzinski PP: Polymorphic variants of folate metabolism genes and the risk of laryngeal cancer. Mol Biol Rep. 2010 Jan;37(1):241-7. doi: 10.1007/s11033-009-9643-y. Epub 2009 Aug 1. [19649727 ]
- Moldt J, Pokorny R, Orth C, Linne U, Geisselbrecht Y, Marahiel MA, Essen LO, Batschauer A: Photoreduction of the folate cofactor in members of the photolyase family. J Biol Chem. 2009 Aug 7;284(32):21670-83. doi: 10.1074/jbc.M109.018697. Epub 2009 Jun 16. [19531478 ]
- Ogwang S, Nguyen HT, Sherman M, Bajaksouzian S, Jacobs MR, Boom WH, Zhang GF, Nguyen L: Bacterial conversion of folinic acid is required for antifolate resistance. J Biol Chem. 2011 Apr 29;286(17):15377-90. doi: 10.1074/jbc.M111.231076. Epub 2011 Mar 3. [21372133 ]
- Watkins D, Rosenblatt DS: Update and new concepts in vitamin responsive disorders of folate transport and metabolism. J Inherit Metab Dis. 2012 Jul;35(4):665-70. doi: 10.1007/s10545-011-9418-1. Epub 2011 Nov 23. [22108709 ]
- Kariluoto S, Edelmann M, Herranen M, Lampi AM, Shmelev A, Salovaara H, Korhola M, Piironen V: Production of folate by bacteria isolated from oat bran. Int J Food Microbiol. 2010 Sep 30;143(1-2):41-7. doi: 10.1016/j.ijfoodmicro.2010.07.026. Epub 2010 Aug 11. [20708290 ]
- Selby CP, Sancar A: The second chromophore in Drosophila photolyase/cryptochrome family photoreceptors. Biochemistry. 2012 Jan 10;51(1):167-71. doi: 10.1021/bi201536w. Epub 2011 Dec 27. [22175817 ]
- Jahansouz H, Scherubel DM, Himes RH: Formylation of tetrahydrofolate by formyl phosphate. FEBS Lett. 1990 Mar 26;262(2):366-8. [2335221 ]
- Tolley M, Bickford L, Clare K, Johann TW: Investigations of amino acids in the ATP binding site of 5,10-methenyltetrahydrofolate synthetase. Protein J. 2012 Aug;31(6):519-28. doi: 10.1007/s10930-012-9428-3. [22773193 ]
- Wijaya IM, Zhang Y, Iwata T, Yamamoto J, Hitomi K, Iwai S, Getzoff ED, Kandori H: Detection of distinct alpha-helical rearrangements of cyclobutane pyrimidine dimer photolyase upon substrate binding by Fourier transform infrared spectroscopy. Biochemistry. 2013 Feb 12;52(6):1019-27. doi: 10.1021/bi3016179. Epub 2013 Jan 30. [23331252 ]
- Upadhyay V, Demmer U, Warkentin E, Moll J, Shima S, Ermler U: Structure and catalytic mechanism of N(5),N(10)-methenyl-tetrahydromethanopterin cyclohydrolase. Biochemistry. 2012 Oct 23;51(42):8435-43. doi: 10.1021/bi300777k. Epub 2012 Oct 8. [23013430 ]
- Kirsch SH, Knapp JP, Herrmann W, Obeid R: Quantification of key folate forms in serum using stable-isotope dilution ultra performance liquid chromatography-tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2010 Jan 1;878(1):68-75. doi: 10.1016/j.jchromb.2009.11.021. Epub . [19959403 ]
- Knock E, Deng L, Krupenko N, Mohan RD, Wu Q, Leclerc D, Gupta S, Elmore CL, Kruger W, Tini M, Rozen R: Susceptibility to intestinal tumorigenesis in folate-deficient mice may be influenced by variation in one-carbon metabolism and DNA repair. J Nutr Biochem. 2011 Nov;22(11):1022-9. doi: 10.1016/j.jnutbio.2010.07.015. Epub 2010 Dec 28. [21193302 ]
|
|---|