|
Record Information |
|---|
| Version |
1.0 |
|---|
| Update Date |
1/22/2018 11:54:54 AM |
|---|
|
Metabolite ID | PAMDB120176 |
|---|
|
Identification |
|---|
| Name: |
S-ureidoglycine |
|---|
| Description: | The zwitterion resulting from the transfer of a proton from the carboxy group to the α-amino group of (S)-2-ureidoglycine. |
|---|
|
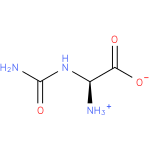
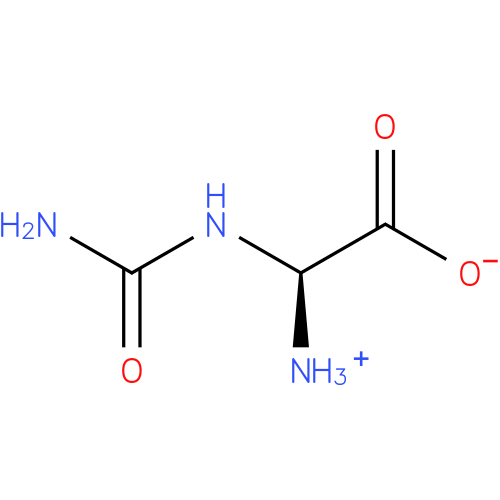
Structure |
|
|---|
| Synonyms: | - (S)-2-azaniumyl-2-(carbamoylamino)acetate
- (S)-2-ureidoglycine
- (S)-ammonio(carbamoylamino)acetate
|
|---|
|
Chemical Formula: |
C3H7N3O3 |
|---|
| Average Molecular Weight: |
133.107 |
|---|
| Monoisotopic Molecular
Weight: |
134.05656 |
|---|
| InChI Key: |
VTFWFHCECSOPSX-SFOWXEAESA-N |
|---|
| InChI: | InChI=1S/C3H7N3O3/c4-1(2(7)8)6-3(5)9/h1H,4H2,(H,7,8)(H3,5,6,9)/t1-/m0/s1 |
|---|
| CAS
number: |
Not Available |
|---|
| IUPAC Name: | (2S)-ammonio(carbamoylamino)ethanoate |
|---|
|
Traditional IUPAC Name: |
Not Available |
|---|
| SMILES: | C(NC(N)=O)([N+])C(=O)[O-] |
|---|
|
Chemical Taxonomy |
|---|
|
Taxonomy Description | This compound belongs to the class of organic compounds known as n-carbamoyl-alpha amino acids. These are compounds containing an alpha amino acid which bears an carbamoyl group at its terminal nitrogen atom. |
|---|
|
Kingdom |
Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
|
Class |
Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
|
Direct Parent |
N-carbamoyl-alpha amino acids |
|---|
| Alternative Parents |
|
|---|
| Substituents |
- N-carbamoyl-alpha-amino acid
- Carboxylic acid salt
- Carbonic acid derivative
- Urea
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Carbonyl group
- Organic zwitterion
- Organic salt
- Organooxygen compound
- Organonitrogen compound
- Hydrocarbon derivative
- Organic oxide
- Organopnictogen compound
- Organic oxygen compound
- Organic nitrogen compound
- Aliphatic acyclic compound
|
|---|
| Molecular Framework |
Aliphatic acyclic compounds |
|---|
| External Descriptors |
Not Available |
|---|
|
Physical Properties |
|---|
| State: |
Not Available |
|---|
| Charge: | 0 |
|---|
|
Melting point: |
Not Available |
|---|
| Experimental Properties: |
Not Available |
|---|
| Predicted Properties |
|
|---|
|
Biological Properties |
|---|
| Cellular Locations: |
Not Available |
|---|
| Reactions: | |
|---|
|
Pathways: |
- superpathway of allantoin degradation in plantsURDEGR-PWY
- allantoin degradation to ureidoglycolate II (ammonia producing)PWY-5698
- allantoin degradation to glyoxylate IIIPWY-5705
|
|---|
|
Spectra |
|---|
| Spectra: |
Not Available |
|---|
|
References |
|---|
| References: |
- Serventi F, Ramazzina I, Lamberto I, Puggioni V, Gatti R, Percudani R (2010)Chemical basis of nitrogen recovery through the ureide pathway: formation and hydrolysis of S-ureidoglycine in plants and bacteria. ACS chemical biology 5, Pubmed: 20038185
|
|---|
| Synthesis Reference: |
Not Available |
|---|
| Material Safety Data Sheet (MSDS) |
Not Available |
|---|
|
Links |
|---|
| External Links: |
|
|---|